Somatic deletions in hereditary breast cancers implicate 13q21 as a putative novel breast cancer susceptibility locus
- PMID: 10944226
- PMCID: PMC16911
- DOI: 10.1073/pnas.97.17.9603
Somatic deletions in hereditary breast cancers implicate 13q21 as a putative novel breast cancer susceptibility locus
Abstract
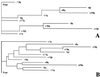
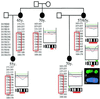
A significant proportion of familial breast cancers cannot be explained by mutations in the BRCA1 or BRCA2 genes. We applied a strategy to identify predisposition loci for breast cancer by using mathematical models to identify early somatic genetic deletions in tumor tissues followed by targeted linkage analysis. Comparative genomic hybridization was used to study 61 breast tumors from 37 breast cancer families with no identified BRCA1 or BRCA2 mutations. Branching and phylogenetic tree models predicted that loss of 13q was one of the earliest genetic events in hereditary cancers. In a Swedish family with five breast cancer cases, all analyzed tumors showed distinct 13q deletions, with the minimal region of loss at 13q21-q22. Genotyping revealed segregation of a shared 13q21 germ-line haplotype in the family. Targeted linkage analysis was carried out in a set of 77 Finnish, Icelandic, and Swedish breast cancer families with no detected BRCA1 and BRCA2 mutations. A maximum parametric two-point logarithm of odds score of 2.76 was obtained for a marker at 13q21 (D13S1308, theta = 0.10). The multipoint logarithm of odds score under heterogeneity was 3.46. The results were further evaluated by simulation to assess the probability of obtaining significant evidence in favor of linkage by chance as well as to take into account the possible influence of the BRCA2 locus, located at a recombination fraction of 0.25 from the new locus. The simulation substantiated the evidence of linkage at D13S1308 (P < 0.0017). The results warrant studies of this putative breast cancer predisposition locus in other populations.
Figures
References
-
- Vehmanen P, Friedman L S, Eerola H, McClure M, Ward B, Sarantaus L, Kainu T, Syrjakoski K, Pyrhonen S, Kallioniemi O-P, et al. Hum Mol Genet. 1997;6:2309–2315. - PubMed
-
- Hemminki A, Tomlinson I, Markie D, Jarvinen H, Sistonen P, Bjorkqvist A M, Knuutila S, Salovaara R, Bodmer W, Shibata D, et al. Nat Genet. 1997;15:87–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

