The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast
- PMID: 10950868
- PMCID: PMC316858
The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast
Abstract
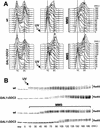
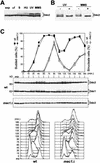
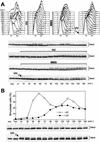
DDC2 is a novel component of the DNA integrity checkpoint pathway, which is required for proper checkpoint response to DNA damage and to incomplete DNA replication. Moreover, Ddc2 overproduction causes sensitivity to DNA-damaging agents and checkpoint defects. Ddc2 physically interacts with Mec1 and undergoes Mec1-dependent phosphorylation both in vitro and in vivo. The phosphorylation of Ddc2 takes place in late S phase and in G(2) phase during an unperturbed cell cycle and is further increased in response to DNA damage. Because Ddc2 phosphorylation does not require any other known tested checkpoint factors but Mec1, the Ddc2-Mec1 complex might respond to the presence of some DNA structures independently of the other known checkpoint proteins. Our findings suggest that Ddc2 may be the functional homolog of Schizosaccharomyces pombe Rad26, strengthening the hypothesis that the mechanisms leading to checkpoint activation are conserved throughout evolution.
Figures



References
-
- Carr AM. Control of cell cycle arrest by the Mec1sc/Rad3sp DNA structure checkpoint pathway. Curr Opin Genet Dev. 1997;7:93–98. - PubMed
-
- Chen G, Yuan SF, Liu W, Xu Y, Trujillo K, Song B, Cong F, Goff SP, Wu Y, Arlinghaus R, et al. Radiation-induced assembly of Rad51 and Rad42 recombination complex requires ATM and c-Abl. J Biol Chem. 1999;274:12748–12752. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
