Transcriptional analyses of antifungal drug resistance in Candida albicans
- PMID: 10952571
- PMCID: PMC90061
- DOI: 10.1128/AAC.44.9.2296-2303.2000
Transcriptional analyses of antifungal drug resistance in Candida albicans
Abstract
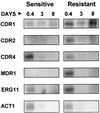
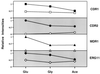
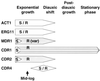
Oral infections with the pathogenic yeast Candida albicans are one of the most frequent and earliest opportunistic infections in human immunodeficiency virus-infected patients. The widespread use of azole antifungal drugs has led to the development of drug-resistant isolates. Several molecular mechanisms that contribute to drug resistance have been identified, including increased mRNA levels for two types of efflux pump genes: the ATP binding cassette transporter CDRs (CDR1 and CDR2) and the major facilitator MDR1. Using Northern blot analyses, the expression patterns of these genes have been determined during logarithmic and stationary phases of cell growth and during growth in different carbon sources in a set of matched susceptible and fluconazole-resistant isolates that have been characterized previously. MDR1, CDR1, and CDR2 are expressed early during logarithmic growth, CDR4 is expressed late during logarithmic growth, and CDR1 is preferentially expressed in stationary-phase cells. There is a small decrease in expression of these genes when the cells are grown in carbon sources other than glucose. While increased mRNA levels of efflux pump genes are commonly associated with azole resistance, the causes of increased mRNA levels have not yet been resolved. Southern blot analysis demonstrates that the increased mRNA levels in these isolates are not the result of gene amplification. Nuclear run-on assays show that MDR1 and CDR mRNAs are transcriptionally overexpressed in the resistant isolate, suggesting that the antifungal drug resistance in this series is associated with the promoter and trans-acting factors of the CDR1, CDR2, and MDR1 genes.
Figures



References
-
- Borst P. Genetic mechanisms of drug resistance. A review. Acta Oncol. 1991;30:87–105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

