Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging
- PMID: 10954568
- PMCID: PMC116378
- DOI: 10.1128/jvi.74.18.8670-8679.2000
Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging
Abstract
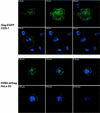
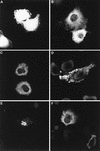
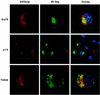
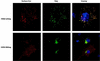
Budding of lentiviruses occurs at the plasma membrane, but the preceding steps involved in particle assembly are poorly understood. Since the Gag polyprotein mediates virion assembly and budding, studies on the localization of Gag within the cell should provide insight into the mechanism of particle assembly. Here, we utilize biochemical fractionation techniques as well as high-resolution confocal imaging of live cells to demonstrate that Gag is localized at the plasma membrane in a striking punctate pattern. Mutation of the N-terminal myristoylation site results in the formation of large cytosolic complexes, whereas mutation of the N-terminal basic residue cluster in the matrix domain redirects the Gag protein to a region partially overlapping the Golgi apparatus. In addition, we show that Gag and Env colocalize at the plasma membrane and that mistargeting of a mutant Gag to the Golgi apparatus alters the pattern of surface expression of Env.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

