Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera
- PMID: 10954570
- PMCID: PMC116380
- DOI: 10.1128/jvi.74.18.8692-8699.2000
Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera
Abstract
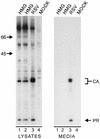
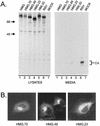
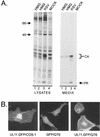
The retroviral Gag protein is capable of directing the production and release of virus-like particles in the absence of all other viral components. Budding normally occurs after Gag is transported to the plasma membrane by its membrane-targeting and -binding (M) domain. In the Rous sarcoma virus (RSV) Gag protein, the M domain is contained within the first 86 amino acids. When M is deleted, membrane association and budding fail to occur. Budding is restored when M is replaced with foreign membrane-binding sequences, such as that of the Src oncoprotein. Moreover, the RSV M domain is capable of targeting heterologous proteins to the plasma membrane. Although the solution structure of the RSV M domain has been determined, the mechanism by which M specifically targets Gag to the plasma membrane rather than to one or more of the large number of internal membrane surfaces (e.g., the Golgi apparatus, endoplasmic reticulum, and nuclear, mitochondrial, or lysosomal membranes) is unknown. To further investigate the requirements for targeting proteins to discrete cellular locations, we have replaced the M domain of RSV with the product of the unique long region 11 (U(L)11) gene of herpes simplex virus type 1. This 96-amino-acid myristylated protein is thought to be involved in virion transport and envelopment at internal membrane sites. When the first 100 amino acids of RSV Gag (including the M domain) were replaced by the entire UL11 sequence, the chimeric protein localized at and budded into the Golgi apparatus rather than being targeted to the plasma membrane. Myristate was found to be required for this specific targeting, as were the first 49 amino acids of UL11, which contain an acidic cluster motif. In addition to shedding new light on UL11, these experiments demonstrate that RSV Gag can be directed to internal cellular membranes and suggest that regions outside of the M domain do not contain a dominant plasma membrane-targeting motif.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

