Transcriptional repression of BRCA1 by aberrant cytosine methylation, histone hypoacetylation and chromatin condensation of the BRCA1 promoter
- PMID: 10954590
- PMCID: PMC110706
- DOI: 10.1093/nar/28.17.3233
Transcriptional repression of BRCA1 by aberrant cytosine methylation, histone hypoacetylation and chromatin condensation of the BRCA1 promoter
Abstract
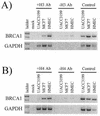
BRCA1 expression is repressed by aberrant cytosine methylation in sporadic breast cancer. We hypothesized that aberrant cytosine methylation of the BRCA1 promoter was associated with the transcriptionally repressive effects of histone hypoacetylation and chromatin condensation. To address this question, we developed an in vitro model of study using normal cells and sporadic breast cancer cells with known levels of BRCA1 transcript to produce a 1.4 kb 5-methylcytosine map of the BRCA1 5' CpG island. While all cell types were densely methylated upstream of -728 relative to BRCA1 transcription start, all normal and BRCA1 expressing cells were non-methylated downstream of -728 suggesting that this region contains the functional BRCA1 5' regulatory region. In contrast, the non-BRCA1 expressing UACC3199 cells were completely methylated at all 75 CpGs. Chromatin immunoprecipitations showed that the UACC3199 cells were hypoacetylated at both histones H3 and H4 in the BRCA1 promoter compared to non-methylated BRCA1 expressing cells. The chromatin of the methylated UACC3199 BRCA1 promoter was inaccessible to DNA-protein interactions. These data indicate that the epigenetic effects of aberrant cytosine methylation, histone hypoacetylation and chromatin condensation act together in a discrete region of the BRCA1 5' CpG island to repress BRCA1 transcription in sporadic breast cancer.
Figures