Accuracy of haplotype frequency estimation for biallelic loci, via the expectation-maximization algorithm for unphased diploid genotype data
- PMID: 10954684
- PMCID: PMC1287896
- DOI: 10.1086/303069
Accuracy of haplotype frequency estimation for biallelic loci, via the expectation-maximization algorithm for unphased diploid genotype data
Abstract
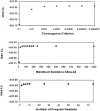
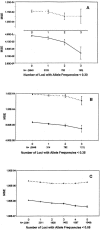
Haplotype analyses have become increasingly common in genetic studies of human disease because of their ability to identify unique chromosomal segments likely to harbor disease-predisposing genes. The study of haplotypes is also used to investigate many population processes, such as migration and immigration rates, linkage-disequilibrium strength, and the relatedness of populations. Unfortunately, many haplotype-analysis methods require phase information that can be difficult to obtain from samples of nonhaploid species. There are, however, strategies for estimating haplotype frequencies from unphased diploid genotype data collected on a sample of individuals that make use of the expectation-maximization (EM) algorithm to overcome the missing phase information. The accuracy of such strategies, compared with other phase-determination methods, must be assessed before their use can be advocated. In this study, we consider and explore sources of error between EM-derived haplotype frequency estimates and their population parameters, noting that much of this error is due to sampling error, which is inherent in all studies, even when phase can be determined. In light of this, we focus on the additional error between haplotype frequencies within a sample data set and EM-derived haplotype frequency estimates incurred by the estimation procedure. We assess the accuracy of haplotype frequency estimation as a function of a number of factors, including sample size, number of loci studied, allele frequencies, and locus-specific allelic departures from Hardy-Weinberg and linkage equilibrium. We point out the relative impacts of sampling error and estimation error, calling attention to the pronounced accuracy of EM estimates once sampling error has been accounted for. We also suggest that many factors that may influence accuracy can be assessed empirically within a data set-a fact that can be used to create "diagnostics" that a user can turn to for assessing potential inaccuracies in estimation.
Figures







References
-
- Clark A (1990) Inference of haplotypes from PCR-amplified samples of diploid populations. Mol Biol Evol 7:111–122 - PubMed
-
- Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927 - PubMed
-
- Hawley M, Kidd K (1995) HAPLO: a program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J Hered 86:409–411 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

