High-resolution quantification of specific mRNA levels in human brain autopsies and biopsies
- PMID: 10958640
- PMCID: PMC310892
- DOI: 10.1101/gr.10.8.1219
High-resolution quantification of specific mRNA levels in human brain autopsies and biopsies
Abstract
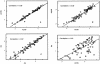
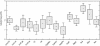
Quantification of mRNA levels in human cortical brain biopsies and autopsies was performed using a fluorogenic 5' nuclease assay. The reproducibility of the assay using replica plates was 97%-99%. Relative quantities of mRNA from 16 different genes were evaluated using a statistical approach based on ANCOVA analysis. Comparison of the relative mRNA levels between two groups of samples with different time postmortem revealed unchanged relative expression levels for most genes. Only CYP26A1 mRNA levels showed a significant decrease with prolonged time postmortem (p = 0.00004). Also, there was a general decrease in measured mRNA levels for all genes in autopsies compared to biopsies; however, on comparing mRNA levels after adjusting with reference genes, no significant differences were found between mRNA levels in autopsies and biopsies. This observation indicates that studies of postmortem material can be performed to reveal the relative in vivo mRNA levels of genes. Power calculations were done to determine the number of individuals necessary to detect differences in mRNA levels of 1.5-fold to tenfold using the strategy described here. This analysis showed that samples from at least 50 individuals per group, patients and controls, are required for high-resolution ( approximately twofold changes) differential expression screenings in the human brain. Experiments done on ten individuals per group will result in a resolution of approximately fivefold changes in expression levels. In general, the sensitivity and resolution of any differential expression study will depend on the sample size used and the between-individual variability of the genes analyzed.
Figures



References
-
- Bailey DS, Bondar A, Furness LM. Pharmacogenomics – it's not just pharmacogenetics. Curr Opin Biotechnol. 1998;9:595–601. - PubMed
-
- Bassett DE, Jr, Eisen MB, Boguski MS. Gene expression informatics–it's all in your mind. Nat Genet. 1999;21:51–55. - PubMed
-
- Benisty S, Boissiere F, Faucheux B, Agid Y, Hirsch EC. trkB messenger RNA expression in normal human brain and in the substantia nigra of parkinsonian patients: An in situ hybridization study. Neuroscience. 1998;86:813–826. - PubMed
-
- Bouchard TJ., Jr Genes, environment, and personality. Science. 1994;264:1700–1701. - PubMed
-
- Cloninger CR, Adolfsson R, Svrakic NM. Mapping genes for human personality [news] Nat Genet. 1996;12:3–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
