Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p
- PMID: 10958662
- PMCID: PMC86166
- DOI: 10.1128/MCB.20.18.6646-6658.2000
Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p
Abstract
In yeast, HOP1 and RED1 are required during meiosis for proper chromosome segregation and the consequent formation of viable spores. Mutations in either HOP1 or RED1 create unique as well as overlapping phenotypes, indicating that the two proteins act alone as well as in concert with each other. To understand which meiotic processes specifically require Red1p-Hop1p hetero-oligomers, a novel genetic screen was used to identify a single-point mutation of RED1, red1-K348E, that separates Hop1p binding from Red1p homo-oligomerization. The Red1-K348E protein is stable, phosphorylated in a manner equivalent to Red1p, and undergoes efficient homo-oligomerization; however, its ability to interact with Hop1p both by two-hybrid and coimmunoprecipitation assays is greatly reduced. Overexpression of HOP1 specifically suppresses red1-K348E, supporting the idea that the only defect in the protein is a reduced affinity for Hop1p. red1-K348E mutants exhibit reduced levels of crossing over and spore viability and fail to undergo chromosome synapsis, thereby implicating a role for Red1p-Hop1p hetero-oligomers in these processes. Furthermore, red1-K348E suppresses the sae2/com1 defects in meiotic progression and sporulation, indicating a previously unknown role for HOP1 in the meiotic recombination checkpoint.
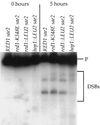
Figures






References
-
- Alani E, Padmore R, Kleckner N. Analysis of wildtype and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. - PubMed
-
- Bai C, Elledge S J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1997;283:141–156. - PubMed
-
- Bailis J M, Roeder G S. Pachytene exit controlled by reversal of Mek1-dependent phosphorylation. Cell. 2000;101:211–221. - PubMed
-
- Bascom-Slack C A, Ross L O, Dawson D S. Chiasmata, crossovers and meiotic chromosome segregation. Adv Genet. 1997;35:253–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
