Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements
- PMID: 10958664
- PMCID: PMC86173
- DOI: 10.1128/MCB.20.18.6668-6676.2000
Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements
Abstract
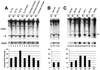
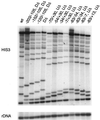
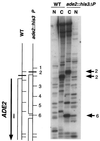
Yeast promoter regions are often more accessible to nuclear proteins than are nonpromoter regions. As assayed by HinfI endonuclease cleavage in living yeast cells, HinfI sites located in the promoters of all seven genes tested were 5- to 20-fold more accessible than sites in adjacent nonpromoter regions. HinfI hypersensitivity within the his3 promoter region is locally determined, since it was observed when this region was translocated to the middle of the ade2 structural gene. Detailed analysis of the his3 promoter indicated that preferential accessibility is not determined by specific elements such as the Gcn4 binding site, poly(dA-dT) sequences, TATA elements, or initiator elements or by transcriptional activity. However, progressive deletion of the promoter region in either direction resulted in a progressive loss of HinfI accessibility. Preferential accessibility is independent of the Swi-Snf chromatin remodeling complex, Gcn5 histone acetylase complexes Ada and SAGA, and Rad6, which ubiquitinates histone H2B. These results suggest that preferential accessibility of the his3 (and presumably other) promoter regions is determined by a general property of the DNA sequence (e.g., base composition or a related feature) rather than by defined sequence elements. The organization of the compact yeast genome into inherently distinct promoter and nonpromoter regions may ensure that transcription factors bind preferentially to appropriate sites in promoters rather than to the excess of irrelevant but equally high-affinity sites in nonpromoter regions.
Figures

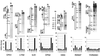




References
-
- Axelrod J D, Majors J. GAL4 disrupts a nucleosome to activate GAL1 transcription in vivo. Genes Dev. 1993;7:857–869. - PubMed
-
- Drew H R, Travers A A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985;186:773–790. - PubMed
-
- Eibel H, Philippsen P. Preferential integration of yeast transposable element Ty into a promoter region. Nature. 1984;307:386–388. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
