Role of the LXCXE binding site in Rb function
- PMID: 10958676
- PMCID: PMC86207
- DOI: 10.1128/MCB.20.18.6799-6805.2000
Role of the LXCXE binding site in Rb function
Abstract
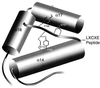
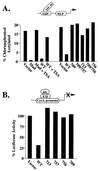
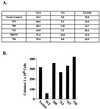
Oncoproteins from DNA tumor viruses such as adenovirus E1a, simian virus 40 T antigen, and human papillomavirus E7 contain an LXCXE sequence, which they use to bind the retinoblastoma protein (Rb) and inhibit its function. Cellular proteins such as histone deacetylases 1 and 2 (HDAC1 and -2) also contain an LXCXE-like sequence, which they use to interact with Rb. The LXCXE binding site in Rb was mutated to assess its role in Rb function. These mutations inhibited binding to HDAC1 and -2, which each contain an LXCXE-like sequence, but had no effect on binding to HDAC3, which lacks an LXCXE-like sequence. Mutation of the LXCXE binding site inhibited active transcriptional repression by Rb and prevented it from effectively repressing the cyclin E and A gene promoters. In contrast, mutations in the LXCXE binding site did not prevent Rb from binding and inactivating E2F. Thus, the LXCXE mutations appear to separate Rb's ability to bind and inactivate E2F from its ability to efficiently recruit HDAC1 and -2 and actively repress transcription. In transient assays, several of the LXCXE binding site mutants caused an increase in the percentage of cells in G(1) by flow cytometry, suggesting that they can arrest cells. However, this effect was transient, as none of the mutants affected cell proliferation in longer-term assays examining bromodeoxyuridine incorporation or colony formation. Our results then suggest that the LXCXE binding site is important for full Rb function. Mutation of the LXCXE binding site does not inhibit binding of the BRG1 ATPase component of the SWI/SNF nucleosome remodeling complex, which has been shown previously to be important for Rb function. Indeed, overexpression of BRG1 and Rb in cells deficient for the proteins led to stable growth inhibition, suggesting a cooperative role for SWI/SNF and the LXCXE binding site in efficient Rb function.
Figures



References
-
- Adams P D, Kaelin W G., Jr The cellular effects of E2F overexpression. Curr Top Microbiol Immunol. 1996;208:79–93. - PubMed
-
- Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. - PubMed
-
- Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
