Identifying a core RNA polymerase surface critical for interactions with a sigma-like specificity factor
- PMID: 10958696
- PMCID: PMC88776
- DOI: 10.1128/MCB.20.18.7013-7023.2000
Identifying a core RNA polymerase surface critical for interactions with a sigma-like specificity factor
Abstract
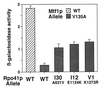
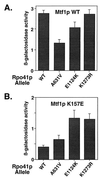
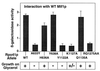
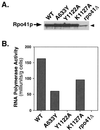
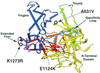
Cyclic interactions occurring between a core RNA polymerase (RNAP) and its initiation factors are critical for transcription initiation, but little is known about subunit interaction. In this work we have identified regions of the single-subunit yeast mitochondrial RNAP (Rpo41p) important for interaction with its sigma-like specificity factor (Mtf1p). Previously we found that the whole folded structure of both polypeptides as well as specific amino acids in at least three regions of Mtf1p are required for interaction. In this work we started with an interaction-defective point mutant in Mtf1p (V135A) and used a two-hybrid selection to isolate suppressing mutations in the core polymerase. We identified suppressors in three separate regions of the RNAP which, when modeled on the structure of the closely related phage T7 RNAP, appear to lie on one surface of the protein. Additional point mutations and biochemical assays were used to confirm the importance of each region for Rpo41p-Mtf1p interactions. Remarkably, two of the three suppressors are found in regions required by T7 RNAP for DNA sequence recognition and promoter melting. Although these essential regions of the phage RNAP are poorly conserved with the mitochondrial RNAPs, they are conserved among the mitochondrial enzymes. The organellar RNAPs appear to use this surface in an alternative way for interactions with their separate sigma-like specificity factor, which, like its bacterial counterpart, provides promoter recognition and DNA melting functions to the holoenzyme.
Figures



References
-
- Arthur T M, Burgess R R. Localization of a sigma70 binding site on the N terminus of the Escherichia coli RNA polymerase beta′ subunit. J Biol Chem. 1998;273:31381–31387. - PubMed
-
- Callaci S, Heyduk E, Heyduk T. Conformational changes of Escherichia coli RNA polymerase sigma70 factor induced by binding to the core enzyme. J Biol Chem. 1998;273:32995–33001. - PubMed
-
- Callaci S, Heyduk E, Heyduk T. Core RNA polymerase from E. coli induces a major change in the domain arrangement of the sigma 70 subunit. Mol Cell. 1999;3:229–238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
