Defective peroxisome membrane synthesis due to mutations in human PEX3 causes Zellweger syndrome, complementation group G
- PMID: 10958759
- PMCID: PMC1287898
- DOI: 10.1086/303071
Defective peroxisome membrane synthesis due to mutations in human PEX3 causes Zellweger syndrome, complementation group G
Abstract
Zellweger cerebro-hepato-renal syndrome is a severe congenital disorder associated with defective peroxisomal biogenesis. At least 23 PEX genes have been reported to be essential for peroxisome biogenesis in various species, indicating the complexity of peroxisomal assembly. Cells from patients with peroxisomal biogenesis disorders have previously been shown to segregate into >/=12 complementation groups. Two patients assigned to complementation group G who had not been linked previously to a specific gene defect were confirmed as displaying a cellular phenotype characterized by a lack of even residual peroxisomal membrane structures. Here we demonstrate that this complementation group is associated with mutations in the PEX3 gene, encoding an integral peroxisomal membrane protein. Homozygous PEX3 mutations, each leading to C-terminal truncation of PEX3, were identified in the two patients, who both suffered from a severe Zellweger syndrome phenotype. One of the mutations involved a single-nucleotide insertion in exon 7, whereas the other was a single-nucleotide substitution eight nucleotides from the normal splice site in the 3' acceptor site of intron 10. Expression of wild-type PEX3 in the mutant cell lines restored peroxisomal biogenesis, whereas transfection of mutated PEX3 cDNA did not. This confirmed that the causative gene had been identified. The observation of peroxisomal formation in the absence of morphologically recognizable peroxisomal membranes challenges the theory that peroxisomes arise exclusively by growth and division from preexisting peroxisomes and establishes PEX3 as a key factor in early human peroxisome synthesis.
Figures
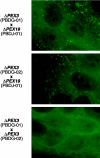


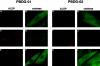

References
Electronic-Database Information
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for PBD [MIM 601539], RCDP [MIM 215100], and PEX3 [MIM 603164])
References
-
- Baerends RJ, Rasmussen SW, Hilbrands RE, van der Heide M, Faber KN, Reuvekamp PT, Kiel JA, Cregg JM, van der Klei IJ, Veenhuis M (1996) The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem 271:8887–8894 - PubMed
-
- Glöckner CJ, Mayerhofer PU, Landgraf P, Muntau AC, Holzinger A, Gerber JK, Kammerer S, Adamski J, Roscher AA (2000) Human adrenoleukodystrophy protein and related peroxisomal ABC transporters interact with the peroxisomal assembly protein PEX19p. Biochem Biophys Res Commun 271:144–150 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

