The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis
- PMID: 10960088
- PMCID: PMC94652
- DOI: 10.1128/JB.182.18.5059-5069.2000
The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis
Abstract
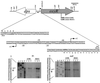
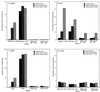
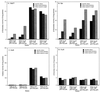
The peroxide response-inducible genes ahpCF, dps, and katB in the obligate anaerobe Bacteroides fragilis are controlled by the redox-sensitive transcriptional activator OxyR. This is the first functional oxidative stress regulator identified and characterized in anaerobic bacteria. oxyR and dps were found to be divergently transcribed, with an overlap in their respective promoter regulatory regions. B. fragilis OxyR and Dps proteins showed high identity to homologues from a closely related anaerobe, Porphyromonas gingivalis. Northern blot analysis revealed that oxyR was expressed as a monocistronic 1-kb mRNA and that dps mRNA was approximately 500 bases in length. dps mRNA was induced over 500-fold by oxidative stress in the parent strain and was constitutively induced in the peroxide-resistant mutant IB263. The constitutive peroxide response in strain IB263 was shown to have resulted from a missense mutation at codon 202 (GAT to GGT) of the oxyR gene [oxyR(Con)] with a predicted D202G substitution in the OxyR protein. Transcriptional fusion analysis revealed that deletion of oxyR abolished the induction of ahpC and katB following treatment with hydrogen peroxide or oxygen exposure. However, dps expression was induced approximately fourfold by oxygen exposure in DeltaoxyR strains but not by hydrogen peroxide. This indicates that dps expression is also under the control of an oxygen-dependent OxyR-independent mechanism. Complementation of DeltaoxyR mutant strains with wild-type oxyR and oxyR(Con) restored the inducible peroxide response and the constitutive response of the ahpCF, katB, and dps genes, respectively. However, overexpression of OxyR abolished the catalase activity but not katB expression, suggesting that higher levels of intracellular OxyR may be involved in other physiological processes. Analysis of oxyR expression in the parents and in DeltaoxyR and overexpressing oxyR strains by Northern blotting and oxyR'::xylB fusions revealed that B. fragilis OxyR does not control its own expression.
Figures


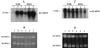
Similar articles
-
Genetic analysis of an important oxidative stress locus in the anaerobe Bacteroides fragilis.Gene. 2003 Oct 16;316:167-75. doi: 10.1016/s0378-1119(03)00759-5. Gene. 2003. PMID: 14563563
-
Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate Anaerobe bacteroides fragilis.J Bacteriol. 1999 Sep;181(18):5701-10. doi: 10.1128/JB.181.18.5701-5710.1999. J Bacteriol. 1999. PMID: 10482511 Free PMC article.
-
Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF.J Bacteriol. 2000 Aug;182(16):4533-44. doi: 10.1128/JB.182.16.4533-4544.2000. J Bacteriol. 2000. PMID: 10913087 Free PMC article.
-
The OxyR regulon.Antonie Van Leeuwenhoek. 1990 Oct;58(3):157-61. doi: 10.1007/BF00548927. Antonie Van Leeuwenhoek. 1990. PMID: 2256675 Review.
-
Regulation of inducible peroxide stress responses.Mol Microbiol. 2002 Jul;45(1):9-15. doi: 10.1046/j.1365-2958.2002.03015.x. Mol Microbiol. 2002. PMID: 12100544 Review.
Cited by
-
Dps and DpsL Mediate Survival In Vitro and In Vivo during the Prolonged Oxidative Stress Response in Bacteroides fragilis.J Bacteriol. 2015 Oct;197(20):3329-38. doi: 10.1128/JB.00342-15. Epub 2015 Aug 10. J Bacteriol. 2015. PMID: 26260459 Free PMC article.
-
Insertional Inactivation of Prevotella intermedia OxyR Results in Reduced Survival with Oxidative Stress and in the Presence of Host Cells.Microorganisms. 2021 Mar 7;9(3):551. doi: 10.3390/microorganisms9030551. Microorganisms. 2021. PMID: 33800047 Free PMC article.
-
Extracellular Vesicles of Porphyromonas gingivalis Disrupt Trophoblast Cell Interaction with Vascular and Immune Cells in an In Vitro Model of Early Placentation.Life (Basel). 2023 Sep 27;13(10):1971. doi: 10.3390/life13101971. Life (Basel). 2023. PMID: 37895353 Free PMC article.
-
Thioredoxins in redox maintenance and survival during oxidative stress of Bacteroides fragilis.J Bacteriol. 2009 May;191(10):3384-91. doi: 10.1128/JB.01665-08. Epub 2009 Mar 13. J Bacteriol. 2009. PMID: 19286811 Free PMC article.
-
Gene expression in Porphyromonas gingivalis after contact with human epithelial cells.Infect Immun. 2005 Apr;73(4):2327-35. doi: 10.1128/IAI.73.4.2327-2335.2005. Infect Immun. 2005. PMID: 15784578 Free PMC article.
References
-
- Almirón M, Link A J, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. - PubMed
-
- Altuvia S, Almirón M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and ςS in stationary phase. Mol Microbiol. 1994;13:265–272. - PubMed
-
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

