Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori
- PMID: 10960091
- PMCID: PMC94655
- DOI: 10.1128/JB.182.18.5082-5090.2000
Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori
Abstract
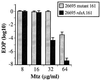
Helicobacter pylori is a human-pathogenic bacterial species that is subdivided geographically, with different genotypes predominating in different parts of the world. Here we test and extend an earlier conclusion that metronidazole (Mtz) resistance is due to mutation in rdxA (HP0954), which encodes a nitroreductase that converts Mtz from prodrug to bactericidal agent. We found that (i) rdxA genes PCR amplified from 50 representative Mtz(r) strains from previously unstudied populations in Asia, South Africa, Europe, and the Americas could, in each case, transform Mtz(s) H. pylori to Mtz(r); (ii) Mtz(r) mutant derivatives of a cultured Mtz(s) strain resulted from mutation in rdxA; and (iii) transformation of Mtz(s) strains with rdxA-null alleles usually resulted in moderate level Mtz resistance (16 microg/ml). However, resistance to higher Mtz levels was common among clinical isolates, a result that implicates at least one additional gene. Expression in Escherichia coli of frxA (HP0642; flavin oxidoreductase), an rdxA paralog, made this normally resistant species Mtz(s), and frxA inactivation enhanced Mtz resistance in rdxA-deficient cells but had little effect on the Mtz susceptibility of rdxA(+) cells. Strains carrying frxA-null and rdxA-null alleles could mutate to even higher resistance, a result implicating one or more additional genes in residual Mtz susceptibility and hyperresistance. We conclude that most Mtz resistance in H. pylori depends on rdxA inactivation, that mutations in frxA can enhance resistance, and that genes that confer Mtz resistance without rdxA inactivation are rare or nonexistent in H. pylori populations.
Figures
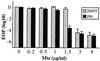
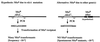
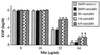
References
-
- Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z J, Suerbaum S, Thompson S A, Van Der Ende A, Van Doorn L J. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. - PubMed
-
- Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. - PubMed
-
- Ausubel F M, et al., editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1998. p. 1.1.1.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

