ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains
- PMID: 10960100
- PMCID: PMC94664
- DOI: 10.1128/JB.182.18.5153-5166.2000
ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains
Abstract
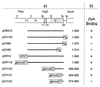
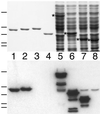
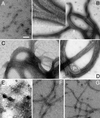
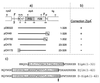
FtsZ and ZipA are essential components of the septal ring apparatus, which mediates cell division in Escherichia coli. FtsZ is a cytoplasmic tubulin-like GTPase that forms protofilament-like homopolymers in vitro. In the cell, the protein assembles into a ring structure at the prospective division site early in the division cycle, and this marks the first recognized event in the assembly of the septal ring. ZipA is an inner membrane protein which is recruited to the nascent septal ring at a very early stage through a direct interaction with FtsZ. Using affinity blotting and protein localization techniques, we have determined which domain on each protein is both sufficient and required for the interaction between the two proteins in vitro as well as in vivo. The results show that ZipA binds to residues confined to the 20 C-terminal amino acids of FtsZ. The FtsZ binding (FZB) domain of ZipA is significantly larger and encompasses the C-terminal 143 residues of ZipA. Significantly, we find that the FZB domain of ZipA is also required and sufficient to induce dramatic bundling of FtsZ protofilaments in vitro. Consistent with the notion that the ability to bind and bundle FtsZ polymers is essential to the function of ZipA, we find that ZipA derivatives lacking an intact FZB domain fail to support cell division in cells depleted for the native protein. Interestingly, ZipA derivatives which do contain an intact FZB domain but which lack the N-terminal membrane anchor or in which this anchor is replaced with the heterologous anchor of the DjlA protein also fail to rescue ZipA(-) cells. Thus, in addition to the C-terminal FZB domain, the N-terminal domain of ZipA is required for ZipA function. Furthermore, the essential properties of the N domain may be more specific than merely acting as a membrane anchor.
Figures







References
-
- Blanar M A, Rutter W J. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. - PubMed
-
- Broome-Smith J K, Spratt B G. A vector for the construction of translational fusions to TEM β-lactamase and the analysis of protein export signals and membrane protein topology. Gene. 1986;49:341–349. - PubMed
-
- Clarke D J, Jacq A, Holland I B. A novel DnaJ-like protein in Escherichia coli inserts into the cytoplasmic membrane with a type III topology. Mol Microbiol. 1996;20:1273–1286. - PubMed
-
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. - PubMed
-
- de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

