The importance of reactant positioning in enzyme catalysis: a hybrid quantum mechanics/molecular mechanics study of a haloalkane dehalogenase
- PMID: 10963662
- PMCID: PMC27632
- DOI: 10.1073/pnas.97.18.9937
The importance of reactant positioning in enzyme catalysis: a hybrid quantum mechanics/molecular mechanics study of a haloalkane dehalogenase
Abstract
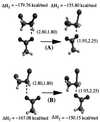
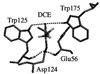
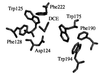
Hybrid quantum mechanics/molecular mechanics calculations using Austin Model 1 system-specific parameters were performed to study the S(N)2 displacement reaction of chloride from 1,2-dichloroethane (DCE) by nucleophilic attack of the carboxylate of acetate in the gas phase and by Asp-124 in the active site of haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. The activation barrier for nucleophilic attack of acetate on DCE depends greatly on the reactants having a geometry resembling that in the enzyme or an optimized gas-phase structure. It was found in the gas-phase calculations that the activation barrier is 9 kcal/mol lower when dihedral constraints are used to restrict the carboxylate nucleophile geometry to that in the enzyme relative to the geometries for the reactants without dihedral constraints. The calculated quantum mechanics/molecular mechanics activation barriers for the enzymatic reaction are 16.2 and 19.4 kcal/mol when the geometry of the reactants is in a near attack conformer from molecular dynamics and in a conformer similar to the crystal structure (DCE is gauche), respectively. This haloalkane dehalogenase lowers the activation barrier for dehalogenation of DCE by 2-4 kcal/mol relative to the single point energies of the enzyme's quantum mechanics atoms in the gas phase. S(N)2 displacements of this sort in water are infinitely slower than in the gas phase. The modest lowering of the activation barrier by the enzyme relative to the reaction in the gas phase is consistent with mutation experiments.
Figures



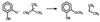
References
-
- Schanstra J P, Kingma J, Janssen D B. J Biol Chem. 1996;271:14747–14753. - PubMed
-
- Verschueren K H G, Seljee F, Rozenboom H J, Kalk K H, Dijkstra B W. Nature (London) 1993;363:693–698. - PubMed
-
- Pries F, Kingma J, Krooshof G H, Jeronimus-Stratingh C M, Bruins A P, Janssen D B. J Biol Chem. 1995;270:10405–10411. - PubMed
-
- Stucki G, Thuer M. Appl Microbiol Biotechnol. 1994;42:167–172. - PubMed
-
- Kennes C, Pries F, Krooshof G H, Bokma E, Kingma J, Janssen D J. Eur J Biochem. 1995;228:403–407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

