Microbial community dynamics during production of the Mexican fermented maize dough pozol
- PMID: 10966374
- PMCID: PMC92204
- DOI: 10.1128/AEM.66.9.3664-3673.2000
Microbial community dynamics during production of the Mexican fermented maize dough pozol
Abstract
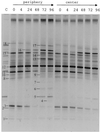
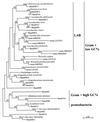
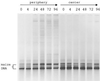
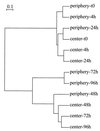
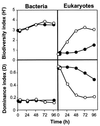
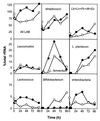
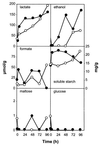
The dynamics of the microbial community responsible for the traditional fermentation of maize in the production of Mexican pozol was investigated by using a polyphasic approach combining (i) microbial enumerations with culture media, (ii) denaturing gradient gel electrophoresis (DGGE) fingerprinting of total community DNA with bacterial and eukaryotic primers and sequencing of partial 16S ribosomal DNA (rDNA) genes, (iii) quantification of rRNAs from dominant microbial taxa by using phylogenetic oligonucleotide probes, and (iv) analysis of sugars and fermentation products. A Streptococcus species dominated the fermentation and accounted for between 25 and 75% of the total flora throughout the process. Results also showed that the initial epiphytic aerobic microflora was replaced in the first 2 days by heterofermentative lactic acid bacteria (LAB), including a close relative of Lactobacillus fermentum, producing lactic acid and ethanol; this heterolactic flora was then progressively replaced by homofermentative LAB (mainly close relatives of L. plantarum, L. casei, and L. delbrueckii) which continued acidification of the maize dough. At the same time, a very diverse community of yeasts and fungi developed, mainly at the periphery of the dough. The analysis of the DGGE patterns obtained with bacterial and eukaryotic primers targeting the 16S and 18S rDNA genes clearly demonstrated that there was a major shift in the community structure after 24 h and that high biodiversity-according to the Shannon-Weaver index-was maintained throughout the process. These results proved that a relatively high number of species, at least six to eight, are needed to perform this traditional lactic acid fermentation. The presence of Bifidobacterium, Enterococcus, and enterobacteria suggests a fecal origin of some important pozol microorganisms. Overall, the results obtained with different culture-dependent or -independent techniques clearly confirmed the importance of developing a polyphasic approach to study the ecology of fermented foods.
Figures


References
-
- Adler-Nissen J, Demain A L. Aeration-controlled formation of acetic acid in heterolactic fermentations. J Ind Microbiol. 1994;13:335–343.
-
- Agati V, Guyot J-P, Morlon-Guyot J, Talamond P, Hounhouigan J. Isolation and characterization of new amylolytic strains of Lactobacillus fermentum from fermented maize doughs (mawè and ogi) from Bénin. J Appl Microbiol. 1998;85:512–520.
-
- Akkermans A D L, Mirza M S, Harmsen H J M, Blok H J, Herron P R, Sessitsch A, Akkermans W M. Molecular ecology of microbes: a review of promises, pitfalls, and true progress. FEMS Microbiol Rev. 1994;15:185–194.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

