Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose
- PMID: 10966429
- PMCID: PMC92259
- DOI: 10.1128/AEM.66.9.4058-4067.2000
Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose
Abstract
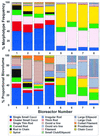
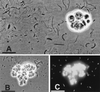
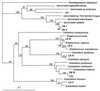
Methanogenic bioreactor communities were used as model ecosystems to evaluate the relationship between functional stability and community structure. Replicated methanogenic bioreactor communities with two different community structures were established. The effect of a substrate loading shock on population dynamics in each microbial community was examined by using morphological analysis, small-subunit (SSU) rRNA oligonucleotide probes, amplified ribosomal DNA (rDNA) restriction analysis (ARDRA), and partial sequencing of SSU rDNA clones. One set of replicated communities, designated the high-spirochete (HS) set, was characterized by good replicability, a high proportion of spiral and short thin rod morphotypes, a dominance of spirochete-related SSU rDNA genes, and a high percentage of Methanosarcina-related SSU rRNA. The second set of communities, designated the low-spirochete (LS) set, was characterized by incomplete replicability, higher morphotype diversity dominated by cocci, a predominance of Streptococcus-related and deeply branching Spirochaetales-related SSU rDNA genes, and a high percentage of Methanosaeta-related SSU rRNA. In the HS communities, glucose perturbation caused a dramatic shift in the relative abundance of fermentative bacteria, with temporary displacement of spirochete-related ribotypes by Eubacterium-related ribotypes, followed by a return to the preperturbation community structure. The LS communities were less perturbed, with Streptococcus-related organisms remaining prevalent after the glucose shock, although changes in the relative abundance of minor members were detected by morphotype analysis. A companion paper demonstrates that the more stable LS communities were less functionally stable than the HS communities (S. A. Hashsham, A. S. Fernandez, S. L. Dollhopf, F. B. Dazzo, R. F. Hickey, J. M. Tiedje, and C. S. Criddle, Appl. Environ. Microbiol. 66:4050-4057, 2000).
Figures


References
-
- Dazzo F B, Petersen M. Applications of computer-assisted image analysis for microscopical studies of the Rhizobium-legume symbiosis. Symbiosis. 1989;7:193–210.
-
- Felsenstein J. Confidence limits of phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

