The multi-PDZ domain protein MUPP1 is a cytoplasmic ligand for the membrane-spanning proteoglycan NG2
- PMID: 10967549
- PMCID: PMC3501957
- DOI: 10.1002/1097-4644(20001101)79:2<213::aid-jcb50>3.0.co;2-g
The multi-PDZ domain protein MUPP1 is a cytoplasmic ligand for the membrane-spanning proteoglycan NG2
Abstract
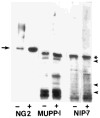
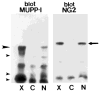
A yeast two-hybrid screen was employed to identify ligands for the cytoplasmic domain of the NG2 chondroitin sulfate proteoglycan. Two overlapping cDNA clones selected in the screen are identical in sequence to a DNA segment coding for the most amino-terminal of the 13 PDZ domains found in the multi-PDZ-protein MUPP1. Antibodies made against recombinant polypeptides representing these two clones (NIP-2 and NIP-7) are reactive with the same 250-kDa molecule recognized by anti-MUPP1 antibodies, confirming the presence of the NIP-2 and NIP-7 sequences in the MUPP1 protein. NIP-2 and NIP-7 GST fusion proteins effectively recognize NG2 in pull-down assays, demonstrating the ability of these polypeptide segments to interact with the intact proteoglycan. The fusion proteins fail to bind NG2 missing the C-terminal half of the cytoplasmic domain, emphasizing the role of the NG2 C-terminus in the interaction with MUPP1. The existence of an NG2/MUPP1 interaction in situ is demonstrated by the ability of NG2 antibodies to co-immunoprecipitate both NG2 and MUPP1 from detergent extracts of cells expressing the two molecules. MUPP1 may serve as a multivalent scaffold that provides a means of linking NG2 with key structural and/or signaling components in the cytoplasm.
Copyright 2000 Wiley-Liss, Inc.
Figures

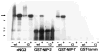
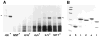
References
-
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl J, editors. Current protocols in molecular biology. New York: John Wiley and Sons, Inc; 1997.
-
- Bartel P, Chien C, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley DA, editor. Cellular interaction in development: a practical approach. Oxford: IRL Press; 1993. pp. 153–179.
-
- Behm F, Smith F, Raimondi S, Pui C, Bernstein I. Human homologue of the rat chondroitin sulfate proteoglycan NG2, detected by monoclonal antibody 7.1, identifies childhood acute lymphoblastic leukemias with t(4;11)(q21;q23) or t(11;19)(q23;13) and MLL gene rearrangements. Blood. 1996;87:1134–1139. - PubMed
-
- Burg M, Grako K, Stallcup W. Expression of the NG2 proteoglycan enhances the growth and metastatic properties of melanoma cells. J Cell Physiol. 1998;177:299–312. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

