The CXXCXXC motif determines the folding, structure and stability of human Ero1-Lalpha
- PMID: 10970843
- PMCID: PMC302061
- DOI: 10.1093/emboj/19.17.4493
The CXXCXXC motif determines the folding, structure and stability of human Ero1-Lalpha
Abstract
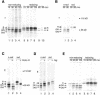
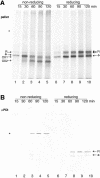
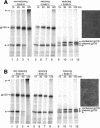
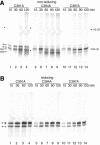
The presence of correctly formed disulfide bonds is crucial to the structure and function of proteins that are synthesized in the endoplasmic reticulum (ER). Disulfide bond formation occurs in the ER owing to the presence of several specialized catalysts and a suitable redox potential. Work in yeast has indicated that the ER resident glycoprotein Ero1p provides oxidizing equivalents to newly synthesized proteins via protein disulfide isomerase (PDI). Here we show that Ero1-Lalpha, the human homolog of Ero1p, exists as a collection of oxidized and reduced forms and covalently binds PDI. We analyzed Ero1-Lalpha cysteine mutants in the presumed active site C(391)VGCFKC(397). Our results demonstrate that this motif is important for protein folding, structural integrity, protein half-life and the stability of the Ero1-Lalpha-PDI complex.
Figures


References
-
- Bader M., Muse,W., Ballou,D.P., Gassner,C. and Bardwell,J.C. (1999) Oxidative protein folding is driven by the electron transport system. Cell, 98, 217–227. - PubMed
-
- Bardwell J.C., McGovern,K. and Beckwith,J. (1991) Identification of a protein required for disulphide bond formation in vivo. Cell, 67, 581–589. - PubMed
-
- Beinert H. and Kiley,P.J. (1999) Fe–S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol., 3, 152–157. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

