Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold
- PMID: 10970852
- PMCID: PMC302050
- DOI: 10.1093/emboj/19.17.4589
Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold
Abstract
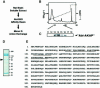
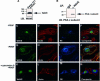
WAVE proteins are members of the Wiskott-Aldrich syndrome protein (WASP) family of scaffolding proteins that coordinate actin reorganization by coupling Rho-related small molecular weight GTPases to the mobilization of the Arp2/3 complex. We identified WAVE-1 in a screen for rat brain A kinase-anchoring proteins (AKAPs), which bind to the SH3 domain of the Abelson tyrosine kinase (Abl). Recombinant WAVE-1 interacts with cAMP-dependent protein kinase (PKA) and Abl kinases when expressed in HEK-293 cells, and both enzymes co-purify with endogenous WAVE from brain extracts. Mapping studies have defined binding sites for each kinase. Competition experiments suggest that the PKA-WAVE-1 interaction may be regulated by actin as the kinase binds to a site overlapping a verprolin homology region, which has been shown to interact with actin. Immunocytochemical analyses in Swiss 3T3 fibroblasts suggest that the WAVE-1 kinase scaffold is assembled dynamically as WAVE, PKA and Abl translocate to sites of actin reorganization in response to platelet-derived growth factor treatment. Thus, we propose a previously unrecognized function for WAVE-1 as an actin-associated scaffolding protein that recruits PKA and Abl.
Figures


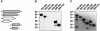

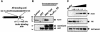



References
-
- Banin S., Gout,I. and Brickell,P. (1999) Interaction between Wiskott–Aldrich syndrome protein (WASP) and the Fyn protein-tyrosine kinase. Mol. Biol. Rep., 26, 173–177. - PubMed
-
- Blanchoin L., Amann,K., Higgs,H., Marchland,J.B., Kaiser,D. and Pollard,T. (2000) Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature, 404, 1007–1011. - PubMed
-
- Bunnell S.C., Henry,P.A., Kolluri,R., Kirchhausen,T., Rickles,R.J. and Berg,L.J. (1996) Identification of Itk/Tsk Src homology 3 domain ligands. J. Biol. Chem., 271, 25646–25656. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

