Regulation of the 26S proteasome by adenovirus E1A
- PMID: 10970867
- PMCID: PMC302057
- DOI: 10.1093/emboj/19.17.4759
Regulation of the 26S proteasome by adenovirus E1A
Abstract
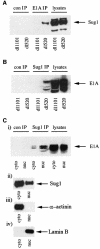
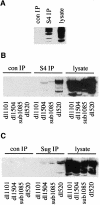
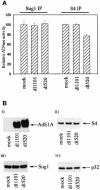
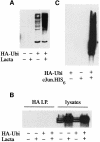
We have identified the N-terminus of adenovirus early region 1A (AdE1A) as a region that can regulate the 26S proteasome. Specifically, in vitro and in vivo co-precipitation studies have revealed that the 19S regulatory components of the proteasome, Sug1 (S8) and S4, bind through amino acids (aa) 4-25 of Ad5 E1A. In vivo expression of wild-type (wt) AdE1A, in contrast to the N-terminal AdE1A mutant that does not bind the proteasome, reduces ATPase activity associated with anti-S4 immunoprecipitates relative to mock-infected cells. This reduction in ATPase activity correlates positively with the ability of wt AdE1A, but not the N-terminal deletion mutant, to significantly reduce the ability of HPV16 E6 to target p53 for ubiquitin-mediated proteasomal degradation. AdE1A/proteasomal complexes are present in both the cytoplasm and the nucleus, suggesting that AdE1A interferes with both nuclear and cytoplasmic proteasomal degradation. We have also demonstrated that wt AdE1A and the N-terminal AdE1A deletion mutant are substrates for proteasomal-mediated degradation. AdE1A degradation is not, however, mediated through ubiquitylation, but is regulated through phosphorylation of residues within a C-terminal PEST region (aa 224-238).
Figures








References
-
- Ait Si Ali S. et al. (1998) Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature, 396, 184–186. - PubMed
-
- Asano K., Vornlocher, H.P., Richter-Cook,N.J., Merrick,W.C., Hinnebusch,A.G. and Hershey,J.W. (1997) Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J. Biol. Chem., 272, 27042–27052. - PubMed
-
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. - PubMed
-
- Baumeister W., Walz,J., Zuhl,F. and Seemuller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. - PubMed
-
- Bayley S.T. and Mymryk,J.S. (1994) Adenovirus E1A proteins and transformation. Int. J. Oncol., 5, 425–444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

