Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain
- PMID: 10973496
- PMCID: PMC27039
- DOI: 10.1073/pnas.190318397
Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain
Abstract
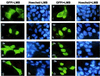
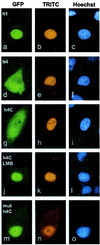
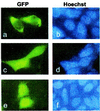
Signal transducer and activator of transcription (Stat) proteins are latent transcription factors that reside in the cytoplasm before activation. On cytokine-induced tyrosine phosphorylation, these molecules dimerize and accumulate transiently in the nucleus. No specific signals mediating these processes have been identified to date. In this report, we examine the nuclear export of Stat1. We find that treatment of cells with the export inhibitor leptomycin B does not affect steady-state localization of Stat1 but impedes nuclear export after IFNgamma-induced nuclear accumulation. We identify a conserved leucine-rich helical segment in the coiled-coil domain of Stat1, which is responsible for the efficient nuclear export of this protein. Mutation of two hallmark leucines within this segment greatly attenuate the back transport of Stat1 in the cytoplasm. When fused to a carrier protein, the Stat1 export sequence can mediate nuclear export after intranuclear microinjection. We show that prolonging the nuclear presence of Stat1 by inhibiting nuclear export reduces the transcriptional response to stimulation with IFNgamma. These data suggest that Stats are actively exported from the nucleus via several separate pathways and link this activity to transcriptional activation.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

