Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription
- PMID: 10973986
- PMCID: PMC2175251
- DOI: 10.1083/jcb.150.5.939
Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription
Abstract
Dentato-rubral and pallido-luysian atrophy (DRPLA) is one of the family of neurodegenerative diseases caused by expansion of a polyglutamine tract. The drpla gene product, atrophin-1, is widely expressed, has no known function or activity, and is found in both the nuclear and cytoplasmic compartments of neurons. Truncated fragments of atrophin-1 accumulate in neuronal nuclei in a transgenic mouse model of DRPLA, and may underlie the disease phenotype. Using the yeast two-hybrid system, we identified ETO/MTG8, a component of nuclear receptor corepressor complexes, as an atrophin-1-interacting protein. When cotransfected into Neuro-2a cells, atrophin-1 and ETO/MTG8 colocalize in discrete nuclear structures that contain endogenous mSin3A and histone deacetylases. These structures are sodium dodecyl sulfate-soluble and associated with the nuclear matrix. Cotransfection of ETO/MTG8 with atrophin-1 recruits atrophin-1 to the nuclear matrix, while atrophin-1 and ETO/MTG8 cofractionate in nuclear matrix preparations from brains of DRPLA transgenic mice. Furthermore, in a cell transfection-based assay, atrophin-1 represses transcription. Together, these results suggest that atrophin-1 associates with nuclear receptor corepressor complexes and is involved in transcriptional regulation. Emerging links between disease-associated polyglutamine proteins, nuclear receptors, translocation-leukemia proteins, and the nuclear matrix may have important repercussions for the pathobiology of this family of neurodegenerative disorders.
Figures



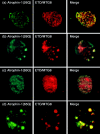
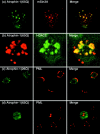
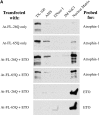



Similar articles
-
Gfi-1 attaches to the nuclear matrix, associates with ETO (MTG8) and histone deacetylase proteins, and represses transcription using a TSA-sensitive mechanism.J Cell Biochem. 2003 Aug 1;89(5):1005-18. doi: 10.1002/jcb.10548. J Cell Biochem. 2003. PMID: 12874834
-
ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain.Mol Cell Biol. 2001 Oct;21(19):6470-83. doi: 10.1128/MCB.21.19.6470-6483.2001. Mol Cell Biol. 2001. PMID: 11533236 Free PMC article.
-
Nuclear localization of a non-caspase truncation product of atrophin-1, with an expanded polyglutamine repeat, increases cellular toxicity.J Biol Chem. 2003 Apr 11;278(15):13047-55. doi: 10.1074/jbc.M211224200. Epub 2002 Dec 2. J Biol Chem. 2003. PMID: 12464607
-
The ETO (MTG8) gene family.Gene. 2003 Jan 16;303:1-10. doi: 10.1016/s0378-1119(02)01172-1. Gene. 2003. PMID: 12559562 Review.
-
Mechanisms of transcriptional repression by the t(8;21)-, t(12;21)-, and inv(16)-encoded fusion proteins.Cancer Chemother Pharmacol. 2001 Aug;48 Suppl 1:S31-4. doi: 10.1007/s002800100302. Cancer Chemother Pharmacol. 2001. PMID: 11587363 Review.
Cited by
-
The corepressor mSin3A regulates phosphorylation-induced activation, intranuclear location, and stability of AML1.Mol Cell Biol. 2004 Feb;24(3):1033-43. doi: 10.1128/MCB.24.3.1033-1043.2004. Mol Cell Biol. 2004. PMID: 14729951 Free PMC article.
-
The expanding role for chromatin and transcription in polyglutamine disease.Curr Opin Genet Dev. 2014 Jun;26:96-104. doi: 10.1016/j.gde.2014.06.008. Epub 2014 Aug 11. Curr Opin Genet Dev. 2014. PMID: 25108806 Free PMC article. Review.
-
Whole-exome sequencing identifies novel candidate predisposition genes for familial polycythemia vera.Hum Genomics. 2017 Apr 20;11(1):6. doi: 10.1186/s40246-017-0102-x. Hum Genomics. 2017. PMID: 28427458 Free PMC article.
-
Consensus paper: pathological mechanisms underlying neurodegeneration in spinocerebellar ataxias.Cerebellum. 2014 Apr;13(2):269-302. doi: 10.1007/s12311-013-0539-y. Cerebellum. 2014. PMID: 24307138 Free PMC article.
-
Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors.Genes Dev. 2006 Mar 1;20(5):525-30. doi: 10.1101/gad.1393506. Epub 2006 Feb 15. Genes Dev. 2006. PMID: 16481466 Free PMC article.
References
-
- Boutell J.M., Thomas P., Neal J.W., Weston V.J., Duce J., Harper P.S., Jones A.L. Aberrant interactions of transcriptional repressor proteins with the Huntington's disease gene product, huntingtin. Hum. Mol. Genet. 1999;8:1647–1655. - PubMed
-
- Cha J.H., Kosinski C.M., Kerner J.A., Alsdorf S.A., Mangiarini L., Davies S.W., Penney J.B., Bates G.P., Young A.B. Altered brain neurotransmitter receptors in transgenic mice expressing a portion of an abnormal human huntington disease gene. Proc. Natl. Acad. Sci. USA. 1998;95:6480–6485. - PMC - PubMed
-
- Chai Y., Koppenhafer S.L., Shoesmith S.J., Perez M.K., Paulson H.L. Evidence for proteasome involvement in polyglutamine diseaselocalization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum. Mol. Genet. 1999;8:673–682. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

