Structure of a conserved receptor domain that regulates kinase activity: the cytoplasmic domain of bacterial taxis receptors
- PMID: 10981636
- PMCID: PMC2902786
- DOI: 10.1016/s0959-440x(00)00115-9
Structure of a conserved receptor domain that regulates kinase activity: the cytoplasmic domain of bacterial taxis receptors
Abstract
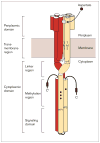
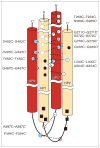
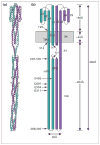
Many bacteria are motile and use a conserved class of transmembrane sensory receptor to regulate cellular taxis toward an optimal living environment. These conserved receptors are typically stimulated by extracellular signals, but also undergo adaptation via covalent modification at specific sites on their cytoplasmic domains. The function of the cytoplasmic domain is to integrate the extracellular and adaptive signals, and to use this integrated information to regulate an associated histidine kinase. The kinase, in turn, triggers a cytoplasmic phosphorylation pathway of the two-component class. The high-resolution structure of a receptor cytoplasmic domain has recently been determined by crystallographic methods and is largely consistent with a structural model independently generated by chemical studies of the domain in the full-length, membrane-bound receptor. These results represent an important step toward a mechanistic understanding of receptor-to-kinase information transfer.
Figures


Similar articles
-
Mechanism of CheA protein kinase activation in receptor signaling complexes.Biochemistry. 1999 May 18;38(20):6651-8. doi: 10.1021/bi982839l. Biochemistry. 1999. PMID: 10350484
-
Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea.J Bacteriol. 2003 Jan;185(1):285-94. doi: 10.1128/JB.185.1.285-294.2003. J Bacteriol. 2003. PMID: 12486065 Free PMC article.
-
Structure, function, and on-off switching of a core unit contact between CheA kinase and CheW adaptor protein in the bacterial chemosensory array: A disulfide mapping and mutagenesis study.Biochemistry. 2013 Nov 5;52(44):7753-65. doi: 10.1021/bi401159k. Epub 2013 Oct 22. Biochemistry. 2013. PMID: 24090207 Free PMC article.
-
Sensor domains of two-component regulatory systems.Curr Opin Microbiol. 2010 Apr;13(2):116-23. doi: 10.1016/j.mib.2010.01.016. Epub 2010 Mar 10. Curr Opin Microbiol. 2010. PMID: 20223701 Free PMC article. Review.
-
Bacterial chemotaxis: a field in motion.Curr Opin Struct Biol. 1995 Dec;5(6):744-51. doi: 10.1016/0959-440x(95)80006-9. Curr Opin Struct Biol. 1995. PMID: 8749361 Review.
Cited by
-
Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching.Biochemistry. 2005 May 31;44(21):7687-95. doi: 10.1021/bi0501479. Biochemistry. 2005. PMID: 15909983 Free PMC article.
-
Evidence that both ligand binding and covalent adaptation drive a two-state equilibrium in the aspartate receptor signaling complex.J Gen Physiol. 2001 Dec;118(6):693-710. doi: 10.1085/jgp.118.6.693. J Gen Physiol. 2001. PMID: 11723162 Free PMC article.
-
Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX.J Bacteriol. 2003 Jan;185(1):89-97. doi: 10.1128/JB.185.1.89-97.2003. J Bacteriol. 2003. PMID: 12486044 Free PMC article.
-
Accessibility of introduced cysteines in chemoreceptor transmembrane helices reveals boundaries interior to bracketing charged residues.Protein Sci. 2004 Jun;13(6):1466-75. doi: 10.1110/ps.04648604. Epub 2004 May 7. Protein Sci. 2004. PMID: 15133159 Free PMC article.
-
Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery.Mol Microbiol. 2006 May;60(4):894-906. doi: 10.1111/j.1365-2958.2006.05145.x. Mol Microbiol. 2006. PMID: 16677301 Free PMC article.
References
-
- Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. The authors describe the crystal structure of the isolated serine receptor cytoplasmic domain, providing the first high-resolution view of this conserved signaling motif, particularly the functionally critical signaling subdomain. - PubMed
-
- Bass RB, Falke JJ. The aspartate receptor cytoplasmic domain: in situ chemical analysis of structure, mechanism and dynamics. Structure. 1999;7:829–840. The chemically derived structure of the dimeric aspartate receptor cytoplasmic domain reveals architectural, dynamical and functional features of the four-helix bundle in the full-length, membrane-bound receptor. - PMC - PubMed
-
- Armitage JP. Bacterial tactic responses. Adv Microb Physiol. 1999;41:229–289. - PubMed
-
- Stock JB, Surette M. Chemotaxis. In: Neidhardt RC, editor. Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology. 2. Washington, DC: ASM Press; 1996. pp. 123–145.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

