Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping
- PMID: 10982322
- PMCID: PMC102074
- DOI: 10.1128/jvi.74.19.8803-8811.2000
Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping
Abstract
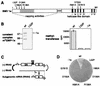
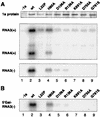
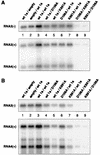
Brome mosaic virus (BMV) encodes two RNA replication proteins: 1a, which contains RNA capping and helicase-like domains, and 2a, which is related to polymerases. BMV 1a and 2a can direct virus-specific RNA replication in the yeast Saccharomyces cerevisiae, which reproduces the known features of BMV replication in plant cells. We constructed single amino acid point mutations at the predicted capping and helicase active sites of 1a and analyzed their effects on BMV RNA3 replication in yeast. The helicase mutants showed no function in any assays used: they were strongly defective in template recruitment for RNA replication, as measured by 1a-induced stabilization of RNA3, and they synthesized no detectable negative-strand or subgenomic RNA. Capping domain mutants divided into two groups. The first exhibited increased template recruitment but nevertheless allowed only low levels of negative-strand and subgenomic mRNA synthesis. The second was strongly defective in template recruitment, made very low levels of negative strands, and made no detectable subgenomes. To distinguish between RNA synthesis and capping defects, we deleted chromosomal gene XRN1, encoding the major exonuclease that degrades uncapped mRNAs. XRN1 deletion suppressed the second but not the first group of capping mutants, allowing synthesis and accumulation of large amounts of uncapped subgenomic mRNAs, thus providing direct evidence for the importance of the viral RNA capping function. The helicase and capping enzyme mutants showed no complementation. Instead, at high levels of expression, a helicase mutant dominantly interfered with the function of the wild-type protein. These results are discussed in relation to the interconnected functions required for different steps of positive-strand RNA virus replication.
Figures




References
-
- Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

