Isolation of a cDNA encoding a granule-bound 152-kilodalton starch-branching enzyme in wheat
- PMID: 10982440
- PMCID: PMC59140
- DOI: 10.1104/pp.124.1.253
Isolation of a cDNA encoding a granule-bound 152-kilodalton starch-branching enzyme in wheat
Abstract
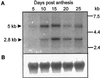
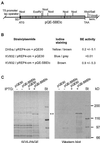
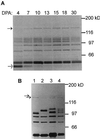
Screening of a wheat (Triticum aestivum) cDNA library for starch-branching enzyme I (SBEI) genes combined with 5'-rapid amplification of cDNA ends resulted in isolation of a 4,563-bp composite cDNA, Sbe1c. Based on sequence alignment to characterized SBEI cDNA clones isolated from plants, the SBEIc predicted from the cDNA sequence was produced with a transit peptide directing the polypeptide into plastids. Furthermore, the predicted mature form of SBEIc was much larger (152 kD) than previously characterized plant SBEI (80-100 kD) and contained a partial duplication of SBEI sequences. The first SBEI domain showed high amino acid similarity to a 74-kD wheat SBEI-like protein that is inactive as a branching enzyme when expressed in Escherichia coli. The second SBEI domain on SBEIc was identical in sequence to a functional 87-kD SBEI produced in the wheat endosperm. Immunoblot analysis of proteins produced in developing wheat kernels demonstrated that the 152-kD SBEIc was, in contrast to the 87- to 88-kD SBEI, preferentially associated with the starch granules. Proteins similar in size and recognized by wheat SBEI antibodies were also present in Triticum monococcum, Triticum tauschii, and Triticum turgidum subsp. durum.
Figures





References
-
- Båga M, Chibbar RN, Kartha KK. Molecular cloning and expression analysis of peroxidase genes from wheat. Plant Mol Biol. 1995;29:647–662. - PubMed
-
- Båga M, Glaze S, Mallard CS, Chibbar RN. A starch branching enzyme gene in wheat produces alternatively spliced transcripts. Plant Mol Biol. 1999;40:1019–1030. - PubMed
-
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J. From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. - PubMed
-
- Bhattacharyya MK, Smith AM, Ellis THN, Hedley C, Martin C. The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell. 1990;60:115–122. - PubMed
-
- Bolt MW, Mahoney PA. High-efficiency blotting of proteins of diverse sizes following sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1997;247:185–192. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

