Specific combinations of zein genes and genetic backgrounds influence the transcription of the heavy-chain zein genes in maize opaque-2 endosperms
- PMID: 10982458
- PMCID: PMC59158
- DOI: 10.1104/pp.124.1.451
Specific combinations of zein genes and genetic backgrounds influence the transcription of the heavy-chain zein genes in maize opaque-2 endosperms
Abstract
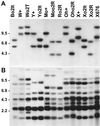
The transcript levels of heavy-chain zein genes (zH1 and zH2) and the occurrence of the zH polypeptides in different opaque-2 (o2) lines were investigated by RNA-blot analyses and by sodium dodecylsulfate-polyacrylamide gel electrophoresis or two-dimensional gel electrophoresis protein fractionations. Four mutant alleles o2R, o2T, o2It, and o2-676 introgressed into different genetic backgrounds (GBs) were considered. The mono-dimensional gel electrophoresis zein pattern can be either conserved or different among the various GBs carrying the same o2 allele. Likewise, in the identical GB carrying different o2 alleles, the zein pattern can be either conserved or differentially affected by the different mutant allele. Zein protein analysis of reciprocal crosses between lines with different o2 alleles or the same o2 showed in some case a more than additive zH pattern in respect to the o2 parent lines. Electrophoretic mobility shift assay approaches, with O2-binding oligonucleotide and endosperm extracts from the above o2 lines, failed to reveal o2-specific retarded band in any of the o2 extracts. The results suggest that the promoter of some zH1 and zH2 contains motif(s) that can respond to factors other than O2.
Figures




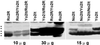
Similar articles
-
Transposable element rbg induces the differential expression of opaque-2 mutant gene in two maize o2 NILs derived from the same inbred line.PLoS One. 2014 Jan 9;9(1):e85159. doi: 10.1371/journal.pone.0085159. eCollection 2014. PLoS One. 2014. PMID: 24416355 Free PMC article.
-
Wild-type opaque2 and defective opaque2 polypeptides form complexes in maize endosperm cells and bind the opaque2-zein target site.Plant Physiol. 2007 Nov;145(3):933-45. doi: 10.1104/pp.107.103606. Epub 2007 Sep 7. Plant Physiol. 2007. PMID: 17827273 Free PMC article.
-
An arginine to lysine substitution in the bZIP domain of an opaque-2 mutant in maize abolishes specific DNA binding.Genes Dev. 1991 Feb;5(2):310-20. doi: 10.1101/gad.5.2.310. Genes Dev. 1991. PMID: 1899843
-
Nitrogen and hormonal responsiveness of the 22 kDa alpha-zein and b-32 genes in maize endosperm is displayed in the absence of the transcriptional regulator Opaque-2.Plant J. 1997 Aug;12(2):281-91. doi: 10.1046/j.1365-313x.1997.12020281.x. Plant J. 1997. PMID: 9301081
-
Manipulation of amino acid balance in maize seeds.Genet Eng (N Y). 1993;15:109-30. doi: 10.1007/978-1-4899-1666-2_5. Genet Eng (N Y). 1993. PMID: 7763837 Review. No abstract available.
Cited by
-
Genetic and molecular understanding for the development of methionine-rich maize: a holistic approach.Front Plant Sci. 2023 Sep 19;14:1249230. doi: 10.3389/fpls.2023.1249230. eCollection 2023. Front Plant Sci. 2023. PMID: 37794928 Free PMC article. Review.
-
Kernel lysine content does not increase in some maize opaque2 mutants.Planta. 2012 Jan;235(1):205-15. doi: 10.1007/s00425-011-1491-z. Epub 2011 Aug 26. Planta. 2012. PMID: 21870098
-
Expressional profiling study revealed unique expressional patterns and dramatic expressional divergence of maize alpha-zein super gene family.Plant Mol Biol. 2009 Apr;69(6):649-59. doi: 10.1007/s11103-008-9444-z. Epub 2008 Dec 28. Plant Mol Biol. 2009. PMID: 19112555
-
Transposable element rbg induces the differential expression of opaque-2 mutant gene in two maize o2 NILs derived from the same inbred line.PLoS One. 2014 Jan 9;9(1):e85159. doi: 10.1371/journal.pone.0085159. eCollection 2014. PLoS One. 2014. PMID: 24416355 Free PMC article.
-
APO2001: A sexy apomixer in como.Plant Cell. 2001 Jul;13(7):1480-91. doi: 10.1105/tpc.13.7.1480. Plant Cell. 2001. PMID: 11449045 Free PMC article. No abstract available.
References
-
- Aukerman MJ, Schmidt RJ. A 168-bp derivative of Suppressor-mutator/Enhancer is responsible for the maize o2-23 mutation. Plant Mol Biol. 1993;21:355–362. - PubMed
-
- Aukerman MJ, Schmidt RJ, Burr B, Burr FA. An arginine to lysine substitution in the bZIP domain of an opaque-2 mutant in maize abolishes specific DNA binding. Genes Dev. 1991;5:310–320. - PubMed
-
- Bernard L, Ciceri P, Viotti A. Molecular analysis of wild-type and mutant alleles at the Opaque-2 regulatory locus reveals different mutations and types of O2 products. Plant Mol Biol. 1994;24:949–959. - PubMed
-
- Bianchi M, Viotti A. DNA methylation and tissue-specific transcription of the storage protein genes of maize. Plant Mol Biol. 1988;11:203–214. - PubMed
-
- Carlini LE, Ketudat M, Parsons RL, Prabhakar S, Schmidt RJ, Guiltiman MJ. The maize EmBP-1 orthologue differentially regulates Opaque2-dependent gene expression in yeast and cultured maize endosperm cells. Plant Mol Biol. 1999;41:339–349. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

