SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories
- PMID: 10982859
- PMCID: PMC110722
- DOI: 10.1093/nar/28.18.3417
SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories
Abstract
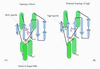
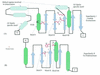
Holliday junction resolvases (HJRs) are key enzymes of DNA recombination. A detailed computer analysis of the structural and evolutionary relationships of HJRs and related nucleases suggests that the HJR function has evolved independently from at least four distinct structural folds, namely RNase H, endonuclease, endonuclease VII-colicin E and RusA. The endonuclease fold, whose structural prototypes are the phage lambda exonuclease, the very short patch repair nuclease (Vsr) and type II restriction enzymes, is shown to encompass by far a greater diversity of nucleases than previously suspected. This fold unifies archaeal HJRs, repair nucleases such as RecB and Vsr, restriction enzymes and a variety of predicted nucleases whose specific activities remain to be determined. Within the RNase H fold a new family of predicted HJRs, which is nearly ubiquitous in bacteria, was discovered, in addition to the previously characterized RuvC family. The proteins of this family, typified by Escherichia coli YqgF, are likely to function as an alternative to RuvC in most bacteria, but could be the principal HJRs in low-GC Gram-positive bacteria and AQUIFEX: Endonuclease VII of phage T4 is shown to serve as a structural template for many nucleases, including MCR:A and other type II restriction enzymes. Together with colicin E7, endonuclease VII defines a distinct metal-dependent nuclease fold. As a result of this analysis, the principal HJRs are now known or confidently predicted for all bacteria and archaea whose genomes have been completely sequenced, with many species encoding multiple potential HJRs. Horizontal gene transfer, lineage-specific gene loss and gene family expansion, and non-orthologous gene displacement seem to have been major forces in the evolution of HJRs and related nucleases. A remarkable case of displacement is seen in the Lyme disease spirochete Borrelia burgdorferi, which does not possess any of the typical HJRs, but instead encodes, in its chromosome and each of the linear plasmids, members of the lambda exonuclease family predicted to function as HJRs. The diversity of HJRs and related nucleases in bacteria and archaea contrasts with their near absence in eukaryotes. The few detected eukaryotic representatives of the endonuclease fold and the RNase H fold have probably been acquired from bacteria via horizontal gene transfer. The identity of the principal HJR(s) involved in recombination in eukaryotes remains uncertain; this function could be performed by topoisomerase IB or by a novel, so far undetected, class of enzymes. Likely HJRs and related nucleases were identified in the genomes of numerous bacterial and eukaryotic DNA viruses. Gene flow between viral and cellular genomes has probably played a major role in the evolution of this class of enzymes. This analysis resulted in the prediction of numerous previously unnoticed nucleases, some of which are likely to be new restriction enzymes.
Figures










References
-
- Holliday R. (1964) Genet. Res., 5, 282–304.
-
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society for Microbiology, Washington, DC.
-
- Lindahl T. and West,S.C. (1995) DNA Repair and Recombination. Chapman & Hall, London, UK.
-
- Smith P.J. and Jones,C. (2000) DNA Recombination and Repair. Oxford University Press, Oxford, UK.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

