Absence of stable intermediates on the folding pathway of barnase
- PMID: 10984513
- PMCID: PMC27103
- DOI: 10.1073/pnas.190265797
Absence of stable intermediates on the folding pathway of barnase
Abstract
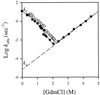
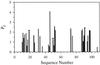
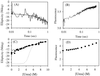
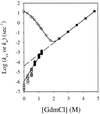
Barnase is one of the few protein models that has been studied extensively for protein folding. Previous studies led to the conclusion that barnase folds through a very stable submillisecond intermediate ( approximately 3 kcal/mol). The structure of this intermediate was characterized intensively by using a protein engineering approach. This intermediate has now been reexamined with three direct and independent methods. (i) Hydrogen exchange experiments show very small protection factors ( approximately 2) for the putative intermediate, indicating a stability of approximately 0.0 kcal/mol. (ii) Denaturant-dependent unfolding of the putative intermediate is noncooperative and indicates a stability less than 0.0 kcal/mol. (iii) The logarithm of the unfolding rate constant of native barnase vs. denaturant concentrations is not linear. Together with the measured rate ("I" to N), this nonlinear behavior accounts for almost all of the protein stability, leaving only about 0.3 kcal/mol that could be attributed to the rapidly formed intermediate. Other observations previously interpreted to support the presence of an intermediate are now known to have alternative explanations. These results cast doubts on the previous conclusions on the nature of the early folding state in barnase and therefore should have important implications in understanding the early folding events of barnase and other proteins in general.
Figures
Similar articles
-
The folding pathway of barnase: the rate-limiting transition state and a hidden intermediate under native conditions.Biochemistry. 2004 Mar 30;43(12):3346-56. doi: 10.1021/bi0362267. Biochemistry. 2004. PMID: 15035606
-
Relationship between the native-state hydrogen exchange and the folding pathways of barnase.Biochemistry. 1999 Oct 26;38(43):14119-24. doi: 10.1021/bi991799y. Biochemistry. 1999. PMID: 10571984
-
Lack of definable nucleation sites in the rate-limiting transition state of barnase under native conditions.J Mol Biol. 2002 Jan 25;315(4):759-70. doi: 10.1006/jmbi.2001.5240. J Mol Biol. 2002. PMID: 11812145
-
The folding of an enzyme. IV. Structure of an intermediate in the refolding of barnase analysed by a protein engineering procedure.J Mol Biol. 1992 Apr 5;224(3):819-35. doi: 10.1016/0022-2836(92)90564-z. J Mol Biol. 1992. PMID: 1569559 Review.
-
The folding of an enzyme. III. Structure of the transition state for unfolding of barnase analysed by a protein engineering procedure.J Mol Biol. 1992 Apr 5;224(3):805-18. doi: 10.1016/0022-2836(92)90563-y. J Mol Biol. 1992. PMID: 1569558 Review.
Cited by
-
Folding subdomains of thioredoxin characterized by native-state hydrogen exchange.Protein Sci. 2003 Aug;12(8):1719-31. doi: 10.1110/ps.0239503. Protein Sci. 2003. PMID: 12876321 Free PMC article.
-
A kinetically significant intermediate in the folding of barnase.Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14121-6. doi: 10.1073/pnas.260502597. Proc Natl Acad Sci U S A. 2000. PMID: 11114199 Free PMC article.
-
Determination of the folding transition states of barnase by using PhiI-value-restrained simulations validated by double mutant PhiIJ-values.Proc Natl Acad Sci U S A. 2005 Aug 30;102(35):12389-94. doi: 10.1073/pnas.0408226102. Epub 2005 Aug 22. Proc Natl Acad Sci U S A. 2005. PMID: 16116084 Free PMC article.
-
Toward correct protein folding potentials.J Biol Phys. 2004 Jun;30(2):171-85. doi: 10.1023/B:JOBP.0000035854.68334.dd. J Biol Phys. 2004. PMID: 23345867 Free PMC article.
-
Mechanical Folding and Unfolding of Protein Barnase at the Single-Molecule Level.Biophys J. 2016 Jan 5;110(1):63-74. doi: 10.1016/j.bpj.2015.11.015. Biophys J. 2016. PMID: 26745410 Free PMC article.
References
-
- Matouschek A, Kellis J T, Jr, Serrano L, Bycroft M, Fersht A R. Nature (London) 1990;346:440–445. - PubMed
-
- Oliveberg M, Fersht A R. Biochemistry. 1996;35:2738–2749. - PubMed
-
- Bycroft M, Matouschek A, Kellis J T, Jr, Serrano L, Fersht A R. Nature (London) 1990;346:488–491. - PubMed
-
- Qi P X, Sosnick T R, Englander S W. Nat Struct Biol. 1998;5:882–887. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

