Unraveling the mechanism of the lactose permease of Escherichia coli
- PMID: 10984523
- PMCID: PMC27091
- DOI: 10.1073/pnas.200351797
Unraveling the mechanism of the lactose permease of Escherichia coli
Abstract
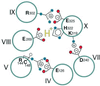
We studied the effect of pH on ligand binding in wild-type lactose permease or mutants in the four residues-Glu-269, Arg-302, His-322, and Glu-325-that are the key participants in H(+) translocation and coupling between sugar and H(+) translocation. Although wild-type permease or mutants in Glu-325 and Arg-302 exhibit marked decreases in affinity at alkaline pH, mutants in either His-322 or Glu-269 do not titrate. The results offer a mechanistic model for lactose/H(+) symport. In the ground state, the permease is protonated, the H(+) is shared between His-322 and Glu-269, Glu-325 is charge-paired with Arg-302, and substrate is bound with high affinity at the outside surface. Substrate binding induces a conformational change that leads to transfer of the H(+) from His-322/Glu-269 to Glu-325 and reorientation of the binding site to the inner surface with a decrease in affinity. Glu-325 then is deprotonated on the inside because of rejuxtaposition with Arg-302. The His-322/Glu-269 complex then is reprotonated from the outside surface to reinitiate the cycle.
Figures

Similar articles
-
Arg-302 facilitates deprotonation of Glu-325 in the transport mechanism of the lactose permease from Escherichiacoli.Proc Natl Acad Sci U S A. 2001 May 22;98(11):6068-73. doi: 10.1073/pnas.111139698. Epub 2001 May 15. Proc Natl Acad Sci U S A. 2001. PMID: 11353849 Free PMC article.
-
Manipulating conformational equilibria in the lactose permease of Escherichia coli.J Mol Biol. 2002 Jan 25;315(4):561-71. doi: 10.1006/jmbi.2001.5289. J Mol Biol. 2002. PMID: 11812130
-
From membrane to molecule to the third amino acid from the left with a membrane transport protein.Q Rev Biophys. 1997 Nov;30(4):333-64. doi: 10.1017/s0033583597003387. Q Rev Biophys. 1997. PMID: 9634651 Review.
-
A molecular mechanism for energy coupling in a membrane transport protein, the lactose permease of Escherichia coli.Proc Natl Acad Sci U S A. 1997 May 27;94(11):5539-43. doi: 10.1073/pnas.94.11.5539. Proc Natl Acad Sci U S A. 1997. PMID: 9159108 Free PMC article.
-
The lactose permease of Escherichia coli: a paradigm for membrane transport proteins.Biochim Biophys Acta. 1992 Jul 17;1101(2):210-3. Biochim Biophys Acta. 1992. PMID: 1633187 Review. No abstract available.
Cited by
-
Structures and General Transport Mechanisms by the Major Facilitator Superfamily (MFS).Chem Rev. 2021 May 12;121(9):5289-5335. doi: 10.1021/acs.chemrev.0c00983. Epub 2021 Apr 22. Chem Rev. 2021. PMID: 33886296 Free PMC article.
-
Protonation and sugar binding to LacY.Proc Natl Acad Sci U S A. 2008 Jul 1;105(26):8896-901. doi: 10.1073/pnas.0803577105. Epub 2008 Jun 20. Proc Natl Acad Sci U S A. 2008. PMID: 18567672 Free PMC article.
-
Cysteine-scanning analysis of helices TM8, TM9a, and TM9b and intervening loops in the YgfO xanthine permease: a carboxyl group is essential at ASP-276.J Biol Chem. 2010 Nov 5;285(45):35011-20. doi: 10.1074/jbc.M110.170415. Epub 2010 Aug 29. J Biol Chem. 2010. PMID: 20802252 Free PMC article.
-
Conservation of residues involved in sugar/H(+) symport by the sucrose permease of Escherichia coli relative to lactose permease.J Mol Biol. 2006 May 12;358(4):1051-9. doi: 10.1016/j.jmb.2006.02.050. Epub 2006 Mar 9. J Mol Biol. 2006. PMID: 16574149 Free PMC article.
-
Lactose permease H+-lactose symporter: mechanical switch or Brownian ratchet?Biophys J. 2007 May 15;92(10):3474-91. doi: 10.1529/biophysj.106.100669. Epub 2007 Feb 26. Biophys J. 2007. PMID: 17325012 Free PMC article.
References
-
- Müller-Hill B. The lac Operon: A Short History of a Genetic Paradigm. Berlin: de Gruyter; 1996.
-
- Kaback H R. J Cell Physiol. 1976;89:575–593. - PubMed
-
- Kaback H R. J Membr Biol. 1983;76:95–112. - PubMed
-
- Frillingos S, Sahin-Tóth M, Wu J, Kaback H R. FASEB J. 1998;12:1281–1299. - PubMed
-
- Kaback H R, Wu J. Acc Chem Res. 1999;32:805–813.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

