An initiation element in the yeast CUP1 promoter is recognized by RNA polymerase II in the absence of TATA box-binding protein if the DNA is negatively supercoiled
- PMID: 10984524
- PMCID: PMC27094
- DOI: 10.1073/pnas.200365097
An initiation element in the yeast CUP1 promoter is recognized by RNA polymerase II in the absence of TATA box-binding protein if the DNA is negatively supercoiled
Abstract
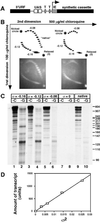
Purified RNA polymerase II initiated transcription from the yeast CUP1 promoter fused to a C-less cassette if the DNA was negatively supercoiled. Relaxed plasmid was not transcribed. Transcription did not require addition of any other transcription factors. TATA box-binding protein (TBP) was not detectable in the polymerase preparation and the TATA box was not required. Deletion analysis of the CUP1 promoter revealed that a 25-bp element containing the initiation region was sufficient for recognition by polymerase. Two transcription start sites were mapped, one of which is identical to one of the two major start sites observed in vivo. Our observations can be accounted for by using a theoretical analysis of the probability of DNA melting within the plasmid as a function of superhelix density: the CUP1 initiation element is intrinsically unstable to superhelical stress, permitting entry of the polymerase, which then scans the DNA to locate the start site. In support of this analysis, the CUP1 promoter was sensitive to mung bean nuclease. These observations and a previous theoretical analysis of yeast genes support the idea that promoters are stress points within the DNA superhelix. The role of transcription factors might be to mark the promoter and to regulate specific melting of promoter DNA.
Figures




Similar articles
-
Identification of the cis-acting DNA sequence elements regulating the transcription of the Saccharomyces cerevisiae gene encoding TBP, the TATA box binding protein.J Biol Chem. 1994 Nov 11;269(45):28335-46. J Biol Chem. 1994. PMID: 7961772
-
A conserved GA element in TATA-less RNA polymerase II promoters.PLoS One. 2011;6(11):e27595. doi: 10.1371/journal.pone.0027595. Epub 2011 Nov 16. PLoS One. 2011. PMID: 22110682 Free PMC article.
-
Mechanistic Differences in Transcription Initiation at TATA-Less and TATA-Containing Promoters.Mol Cell Biol. 2017 Dec 13;38(1):e00448-17. doi: 10.1128/MCB.00448-17. Print 2018 Jan 1. Mol Cell Biol. 2017. PMID: 29038161 Free PMC article.
-
The role of general initiation factors in transcription by RNA polymerase II.Trends Biochem Sci. 1996 Sep;21(9):327-35. Trends Biochem Sci. 1996. PMID: 8870495 Review.
-
The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future.Cold Spring Harb Symp Quant Biol. 1998;63:553-61. doi: 10.1101/sqb.1998.63.553. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384320 Review. No abstract available.
Cited by
-
DNA dynamics play a role as a basal transcription factor in the positioning and regulation of gene transcription initiation.Nucleic Acids Res. 2010 Apr;38(6):1790-5. doi: 10.1093/nar/gkp1084. Epub 2009 Dec 17. Nucleic Acids Res. 2010. PMID: 20019064 Free PMC article.
-
Large-scale structural analysis of the core promoter in mammalian and plant genomes.Nucleic Acids Res. 2005 Jul 27;33(13):4255-64. doi: 10.1093/nar/gki737. Print 2005. Nucleic Acids Res. 2005. PMID: 16049029 Free PMC article.
-
Stress-induced DNA duplex destabilization (SIDD) in the E. coli genome: SIDD sites are closely associated with promoters.Genome Res. 2004 Aug;14(8):1575-84. doi: 10.1101/gr.2080004. Genome Res. 2004. PMID: 15289476 Free PMC article.
-
Toward a detailed description of the thermally induced dynamics of the core promoter.PLoS Comput Biol. 2009 Mar;5(3):e1000313. doi: 10.1371/journal.pcbi.1000313. Epub 2009 Mar 13. PLoS Comput Biol. 2009. PMID: 19282962 Free PMC article.
-
Unravelling the Role of the F55 Regulator in the Transition from Lysogeny to UV Induction of Sulfolobus Spindle-Shaped Virus 1.J Virol. 2015 Jun;89(12):6453-61. doi: 10.1128/JVI.00363-15. Epub 2015 Apr 15. J Virol. 2015. PMID: 25878101 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

