Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori
- PMID: 10986230
- PMCID: PMC110970
- DOI: 10.1128/JB.182.19.5300-5308.2000
Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori
Abstract
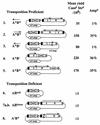
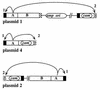
A search by subtractive hybridization for sequences present in only certain strains of Helicobacter pylori led to the discovery of a 2-kb transposable element to be called IS607, which further PCR and hybridization tests indicated was present in about one-fifth of H. pylori strains worldwide. IS607 contained two open reading frames (ORFs) of possibly different phylogenetic origin. One ORF (orfB) exhibited protein-level homology to one of two putative transposase genes found in several other chimeric elements including IS605 (also of H. pylori) and IS1535 (of Mycobacterium tuberculosis). The second IS607 gene (orfA) was unrelated to the second gene of IS605 and might possibly be chimeric itself: it exhibited protein-level homology to merR bacterial regulatory genes in the first approximately 50 codons and homology to the second gene of IS1535 (annotated as "resolvase," apparently due to a weak short recombinase motif) in the remaining three-fourths of its length. IS607 was found to transpose in Escherichia coli, and analyses of sequences of IS607-target DNA junctions in H. pylori and E. coli indicated that it inserted either next to or between adjacent GG nucleotides, and generated either a 2-bp or a 0-bp target sequence duplication, respectively. Mutational tests showed that its transposition in E. coli required orfA but not orfB, suggesting that OrfA protein may represent a new, previously unrecognized, family of bacterial transposases.
Figures



References
-
- Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K A, editors. Current protocols in molecular biology, suppl. 27 CPMB. New York, N.Y: Greene Publishing and Wiley Interscience; 1994. p. 2.4.1.
-
- Berg D E, Gilman R H, Lelwala-Guruge J, Srivastava K, Valdez Y, Watanabe J, Miyagi J, Akopyants N S, Ramirez-Ramos A, Yoshiwara T H, Recavarren S, Leon-Barua R. Helicobacter pyloripopulations in individual Peruvian patients. Clin Infect Dis. 1997;25:996–1002. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

