Molecular analyses of a putative CTXphi precursor and evidence for independent acquisition of distinct CTX(phi)s by toxigenic Vibrio cholerae
- PMID: 10986258
- PMCID: PMC110998
- DOI: 10.1128/JB.182.19.5530-5538.2000
Molecular analyses of a putative CTXphi precursor and evidence for independent acquisition of distinct CTX(phi)s by toxigenic Vibrio cholerae
Abstract
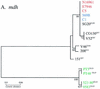
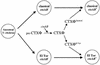
The genes encoding cholera toxin (ctxA and ctxB) are encoded in the genome of CTXphi, a filamentous phage that infects Vibrio cholerae. To study the evolutionary history of CTXphi, we examined genome diversity in CTX(phi)s derived from a variety of epidemic and nonepidemic Vibrio sp. natural isolates. Among these were three V. cholerae strains that contained CTX prophage sequences but not the ctxA and ctxB genes. These prophages each gave rise to a plasmid form whose genomic organization was very similar to that of the CTXphi replicative form, with the exception of missing ctxAB. Sequence analysis of these three plasmids revealed that they lacked the upstream control region normally found 5' of ctxA, as well as the ctxAB promoter region and coding sequences. These findings are consistent with the hypothesis that a CTXphi precursor that lacked ctxAB simultaneously acquired the toxin genes and their regulatory sequences. To assess the evolutionary relationships among additional CTX(phi)s, two CTXphi-encoded genes, orfU and zot, were sequenced from 13 V. cholerae and 4 V. mimicus isolates. Comparative nucleotide sequence analyses revealed that the CTX(phi)s derived from classical and El Tor V. cholerae isolates comprise two distinct lineages within otherwise nearly identical chromosomal backgrounds (based on mdh sequences). These findings suggest that nontoxigenic precursors of the two V. cholerae O1 biotypes independently acquired distinct CTX(phi)s.
Figures







References
-
- Barua B. History of cholera. In: Barua D, Greenough III W B, editors. Cholera. New York, N.Y: Plenum Publishing Corporation; 1992. pp. 1–36.
-
- Bishai W R, Murphy J R. Bacteriophage gene products that cause human disease. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Press; 1988. pp. 683–724.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

