Yersinia enterocolitica ClpB affects levels of invasin and motility
- PMID: 10986262
- PMCID: PMC111002
- DOI: 10.1128/JB.182.19.5563-5571.2000
Yersinia enterocolitica ClpB affects levels of invasin and motility
Abstract
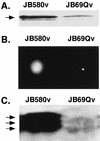
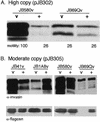
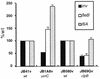
Expression of the Yersinia enterocolitica inv gene is dependent on growth phase and temperature. inv is maximally expressed at 23 degrees C in late-exponential- to early-stationary-phase cultures. We previously reported the isolation of a Y. enterocolitica mutant (JB1A8v) that shows a decrease in invasin levels yet is hypermotile when grown at 23 degrees C. JB1A8v has a transposon insertion within uvrC. Described here is the isolation and characterization of a clone that suppresses these mutant phenotypes of the uvrC mutant JB1A8v. This suppressing clone encodes ClpB (a Clp ATPase homologue). The Y. enterocolitica ClpB homologue is 30 to 40% identical to the ClpB proteins from various bacteria but is 80% identical to one of the two ClpB homologues of Yersinia pestis. A clpB::TnMax2 insertion mutant (JB69Qv) was constructed and determined to be deficient in invasin production and nonmotile when grown at 23 degrees C. Analysis of inv and fleB (flagellin gene) transcript levels in JB69Qv suggested that ClpB has both transcriptional and posttranscriptional effects. In contrast, a clpB null mutant, BY1v, had no effect on invasin levels or motility. A model accounting for these observations is presented.
Figures



References
-
- Bates P F, Swift R A. Double cos site vectors: simplified cosmid cloning. Gene. 1983;26:137–146. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

