Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein
- PMID: 10992471
- PMCID: PMC101523
- DOI: 10.1128/IAI.68.10.5679-5689.2000
Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein
Abstract
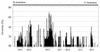
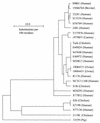
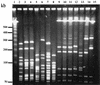
The major outer membrane protein (MOMP), a putative porin and a multifunction surface protein of Campylobacter jejuni, may play an important role in the adaptation of the organism to various host environments. To begin to dissect the biological functions and antigenic features of this protein, the gene (designated cmp) encoding MOMP was identified and characterized from 22 strains of C. jejuni and one strain of C. coli. It was shown that the single-copy cmp locus encoded a protein with characteristics of bacterial outer membrane proteins. Prediction from deduced amino acid sequences suggested that each MOMP subunit consisted of 18 beta-strands connected by short periplasmic turns and long irregular external loops. Alignment of the amino acid sequences of MOMP from different strains indicated that there were seven localized variable regions dispersed among highly conserved sequences. The variable regions were located in the putative external loop structures, while the predicted beta-strands were formed by conserved sequences. The sequence homology of cmp appeared to reflect the phylogenetic proximity of C. jejuni strains, since strains with identical cmp sequences had indistinguishable or closely related macrorestriction fragment patterns. Using recombinant MOMP and antibodies recognizing linear or conformational epitopes of the protein, it was demonstrated that the surface-exposed epitopes of MOMP were predominantly conformational in nature. These findings are instrumental in the design of MOMP-based diagnostic tools and vaccines.
Figures
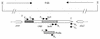



References
-
- Amako K, Baba N, Suzuki N, Wai S N, Umeda A. A structural analysis of the regularly arranged porin on the outer membrane of Campylobacter jejuni based on correlation averaging. Microbiol Immunol. 1997;41:855–859. - PubMed
-
- Amako K, Wai S N, Umeda A, Shigematsu M, Takade A. Electron microscopy of the major outer membrane protein of Campylobacter jejuni. Microbiol Immunol. 1996;40:749–754. - PubMed
-
- Bacon D J, Johnson W M, Rodgers F G. Identification and characterisation of a cytotoxic porin-lipopolysaccharide complex from Campylobacter jejuni. J Med Microbiol. 1999;48:139–148. - PubMed
-
- Black R E, Levine M M, Clements M L, Hughes T P, Blaser M J. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157:472–479. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

