Elucidating the mechanism of familial amyloidosis- Finnish type: NMR studies of human gelsolin domain 2
- PMID: 10995458
- PMCID: PMC27087
- DOI: 10.1073/pnas.180310097
Elucidating the mechanism of familial amyloidosis- Finnish type: NMR studies of human gelsolin domain 2
Abstract
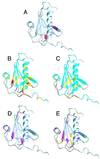
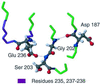
Familial amyloidosis-Finnish type (FAF) results from a single mutation at residue 187 (D187N or D187Y) within domain 2 of the actin-regulating protein gelsolin. The mutation somehow allows a masked cleavage site to be exposed, leading to the first step in the formation of an amyloidogenic fragment. We have performed NMR experiments investigating structural and dynamic changes between wild-type (WT) and D187N gelsolin domain 2 (D2). On mutation, no significant structural or dynamic changes occur at or near the cleavage site. Areas in conformational exchange are observed between beta-strand 4 and alpha-helix 1 and within the loop region following beta-strand 5. Chemical shift differences are noted along the face of alpha-helix 1 that packs onto the beta-sheet, suggesting an altered conformation. Conformational changes within these areas can have an effect on actin binding and may explain why D187N gelsolin is inactive. [(1)H-(15)N] nuclear Overhauser effect and chemical shift data suggest that the C-terminal tail of D187N gelsolin D2 is less structured than WT by up to six residues. In the crystal structure of equine gelsolin, the C-terminal tail of D2 lies across a large cleft between domains 1 and 2 where the masked cleavage site sits. We propose that the D187N mutation destabilizes the C-terminal tail of D2 resulting in a more exposed cleavage site leading to the first proteolysis step in the formation of the amyloidogenic fragment.
Figures


Similar articles
-
Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.Biochim Biophys Acta Mol Basis Dis. 2019 Mar 1;1865(3):648-660. doi: 10.1016/j.bbadis.2019.01.010. Epub 2019 Jan 6. Biochim Biophys Acta Mol Basis Dis. 2019. PMID: 30625383
-
Loss of a metal-binding site in gelsolin leads to familial amyloidosis-Finnish type.Nat Struct Biol. 2002 Feb;9(2):112-6. doi: 10.1038/nsb745. Nat Struct Biol. 2002. PMID: 11753432
-
Gelsolin domain 2 Ca2+ affinity determines susceptibility to furin proteolysis and familial amyloidosis of finnish type.J Mol Biol. 2003 Nov 14;334(1):119-27. doi: 10.1016/j.jmb.2003.09.029. J Mol Biol. 2003. PMID: 14596804
-
Gelsolin amyloidosis: genetics, biochemistry, pathology and possible strategies for therapeutic intervention.Crit Rev Biochem Mol Biol. 2012 May-Jun;47(3):282-96. doi: 10.3109/10409238.2012.661401. Epub 2012 Feb 24. Crit Rev Biochem Mol Biol. 2012. PMID: 22360545 Free PMC article. Review.
-
Ca2+ binding protects against gelsolin amyloidosis.Biochem Biophys Res Commun. 2004 Oct 1;322(4):1105-10. doi: 10.1016/j.bbrc.2004.07.125. Biochem Biophys Res Commun. 2004. PMID: 15336957 Review.
Cited by
-
The disintegration of a molecule: the role of gelsolin in FAF, familial amyloidosis (Finnish type).Proc Natl Acad Sci U S A. 2001 Feb 27;98(5):2117-8. doi: 10.1073/pnas.051635098. Proc Natl Acad Sci U S A. 2001. PMID: 11226199 Free PMC article. No abstract available.
-
The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.Eur Biophys J. 2020 Jan;49(1):11-19. doi: 10.1007/s00249-019-01409-9. Epub 2019 Nov 13. Eur Biophys J. 2020. PMID: 31724080
-
Destabilization of Ca2+-free gelsolin may not be responsible for proteolysis in Familial Amyloidosis of Finnish Type.Proc Natl Acad Sci U S A. 2001 Feb 27;98(5):2334-9. doi: 10.1073/pnas.041452598. Epub 2001 Feb 20. Proc Natl Acad Sci U S A. 2001. PMID: 11226240 Free PMC article.
-
Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.Sci Rep. 2016 Sep 16;6:33463. doi: 10.1038/srep33463. Sci Rep. 2016. PMID: 27633054 Free PMC article.
-
Regulatory role of the second gelsolin-like domain of Caenorhabditis elegans gelsolin-like protein 1 (GSNL-1) in its calcium-dependent conformation and actin-regulatory activities.Cytoskeleton (Hoboken). 2013 Apr;70(4):228-39. doi: 10.1002/cm.21103. Epub 2013 Mar 21. Cytoskeleton (Hoboken). 2013. PMID: 23475707 Free PMC article.
References
-
- Thomas P J, Qu B H, Pedersen P L. Trends Biochem Sci. 1995;20:456–459. - PubMed
-
- Kelly J W. Curr Opin Struct Biol. 1996;6:11–17. - PubMed
-
- Kelly J W. Structure (London) 1997;5:595–600. - PubMed
-
- Kelly J W. Curr Opin Struct Biol. 1998;8:101–106. - PubMed
-
- Blake C, Serpell L. Structure (London) 1996;4:989–998. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

