Biochemical characterization of rotavirus receptors in MA104 cells
- PMID: 11000204
- PMCID: PMC112364
- DOI: 10.1128/jvi.74.20.9362-9371.2000
Biochemical characterization of rotavirus receptors in MA104 cells
Abstract
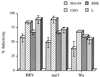
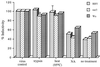
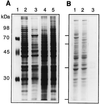
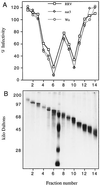
We have tested the effect of metabolic inhibitors, membrane cholesterol depletion, and detergent extraction of cell surface molecules on the susceptibility of MA104 cells to infection by rotaviruses. Treatment of cells with tunicamycin, an inhibitor of protein N glycosylation, blocked the infectivity of the SA-dependent rotavirus RRV and its SA-independent variant nar3 by about 50%, while the inhibition of O glycosylation had no effect. The inhibitor of glycolipid biosynthesis d, l-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol (PDMP) blocked the infectivity of RRV, nar3, and the human rotavirus strain Wa by about 70%. Sequestration of cholesterol from the cell membrane with beta-cyclodextrin reduced the infectivity of the three viruses by more than 90%. The involvement of N-glycoproteins, glycolipids, and cholesterol in rotavirus infection suggests that the virus receptor(s) might be forming part of lipid microdomains in the cell membrane. MA104 cells incubated with the nonionic detergent octyl-beta-glucoside (OG) showed a ca. 60% reduction in their ability to bind rotaviruses, the same degree to which they became refractory to infection, suggesting that OG extracts the potential virus receptor(s) from the cell surface. Accordingly, when preincubated with the viruses, the OG extract inhibited the virus infectivity by more than 95%. This inhibition was abolished when the extract was treated with either proteases or heat but not when it was treated with neuraminidase, indicating the protein nature of the inhibitor. Two protein fractions of around 57 and 75 kDa were isolated from the extract, and these fractions were shown to have rotavirus-blocking activity. Also, antibodies to these fractions efficiently inhibited the infectivity of the viruses in untreated as well as in neuraminidase-treated cells. Five individual protein bands of 30, 45, 57, 75, and 110 kDa, which exhibited virus-blocking activity, were finally isolated from the OG extract. These proteins are good candidates to function as rotavirus receptors.
Figures



Similar articles
-
Non-lytic extraction and characterisation of receptors for multiple strains of rotavirus.Arch Virol. 2001 Jul;146(7):1307-23. doi: 10.1007/s007050170093. Arch Virol. 2001. PMID: 11556708
-
Entry of rotaviruses is a multistep process.Virology. 1999 Oct 25;263(2):450-9. doi: 10.1006/viro.1999.9976. Virology. 1999. PMID: 10544117
-
The VP5 domain of VP4 can mediate attachment of rotaviruses to cells.J Virol. 2000 Jan;74(2):593-9. doi: 10.1128/jvi.74.2.593-599.2000. J Virol. 2000. PMID: 10623720 Free PMC article.
-
Molecular biology of rotavirus cell entry.Arch Med Res. 2002 Jul-Aug;33(4):356-61. doi: 10.1016/s0188-4409(02)00374-0. Arch Med Res. 2002. PMID: 12234525 Review.
-
Early events of rotavirus infection: the search for the receptor(s).Novartis Found Symp. 2001;238:47-60; discussion 60-3. doi: 10.1002/0470846534.ch4. Novartis Found Symp. 2001. PMID: 11444034 Review.
Cited by
-
Function of membrane rafts in viral lifecycles and host cellular response.Biochem Res Int. 2011;2011:245090. doi: 10.1155/2011/245090. Epub 2011 Dec 7. Biochem Res Int. 2011. PMID: 22191032 Free PMC article.
-
Pathogenesis of rotavirus diarrhea.Microbes Infect. 2001 Nov;3(13):1145-56. doi: 10.1016/s1286-4579(01)01475-7. Microbes Infect. 2001. PMID: 11709295 Free PMC article. Review.
-
VP7 mediates the interaction of rotaviruses with integrin alphavbeta3 through a novel integrin-binding site.J Virol. 2004 Oct;78(20):10839-47. doi: 10.1128/JVI.78.20.10839-10847.2004. J Virol. 2004. PMID: 15452204 Free PMC article.
-
The peptide-binding and ATPase domains of recombinant hsc70 are required to interact with rotavirus and reduce its infectivity.J Virol. 2006 Apr;80(7):3322-31. doi: 10.1128/JVI.80.7.3322-3331.2006. J Virol. 2006. PMID: 16537599 Free PMC article.
-
An N-linked glycoprotein with alpha(2,3)-linked sialic acid is a receptor for BK virus.J Virol. 2005 Nov;79(22):14442-5. doi: 10.1128/JVI.79.22.14442-14445.2005. J Virol. 2005. PMID: 16254379 Free PMC article.
References
-
- Basak S, Turner H, Parr S. Identification of a 40- to 42-kDa attachment polypeptide for canine parvovirus in A72 cells. Virology. 1994;205:7–16. - PubMed
-
- Bass D M, Mackow E R, Greenberg H B. Identification and partial characterization of a rhesus rotavirus binding glycoprotein on murine enterocytes. Virology. 1991;183:602–610. - PubMed
-
- Beisner B, Kool D, Marich A, Holmes I H. Characterisation of G serotype dependent non-antibody inhibitors of rotavirus in normal mouse serum. Arch Virol. 1998;143:1277–1294. - PubMed
-
- Brockhausen I, Moller G, Pollex-Kruger A, Rutz V, Paulsen H, Matta K L. Control of O-glycan synthesis: specificity and inhibition of O-glycan core 1 UDP-galactose:N-acetylgalactosamine-alpha-R-beta-3-galactosyltransferase from rat liver. Biochem Cell Biol. 1992;70:99–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

