Transcription and RNA editing in a soluble in vitro system from Physarum mitochondria
- PMID: 11000260
- PMCID: PMC110774
- DOI: 10.1093/nar/28.19.3695
Transcription and RNA editing in a soluble in vitro system from Physarum mitochondria
Abstract
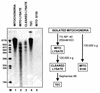
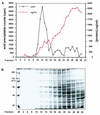
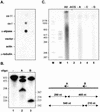
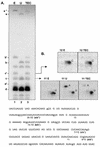
The dissection of RNA editing mechanisms in PHYSARUM: mitochondria has been hindered by the absence of a soluble in vitro system. Based on our studies in isolated mitochondria, insertion of non-encoded nucleotides into PHYSARUM: mitochondrial RNAs is closely linked to transcription. Here we have fractionated mitochondrial lysates, enriching for run-on RNA synthesis, and find that editing activity co-fractionates with pre-formed transcription elongation complexes. The establishment of this soluble transcription-editing system allows access to the components of the editing machinery and permits manipulation of transcription and editing substrates. Thus, the availability of this system provides, for the first time, a means of investigating roles for cis-acting elements, trans-acting factors and nucleotide requirements for the insertion of non-encoded nucleotides into PHYSARUM: mitochondrial RNAs. This methodology should also be broadly applicable to the study of RNA processing and editing mechanisms in a wide range of mitochondrial systems.
Figures
Similar articles
-
Non-templated addition of nucleotides to the 3' end of nascent RNA during RNA editing in Physarum.EMBO J. 2001 Mar 15;20(6):1405-14. doi: 10.1093/emboj/20.6.1405. EMBO J. 2001. PMID: 11250906 Free PMC article.
-
Insertional editing in isolated Physarum mitochondria is linked to RNA synthesis.RNA. 1997 Aug;3(8):821-37. RNA. 1997. PMID: 9257642 Free PMC article.
-
Accurate and efficient insertional RNA editing in isolated Physarum mitochondria.RNA. 1995 Sep;1(7):681-91. RNA. 1995. PMID: 7585253 Free PMC article.
-
[RNA editing in different genetic systems].Zh Obshch Biol. 2004 Jan-Feb;65(1):52-73. Zh Obshch Biol. 2004. PMID: 15032065 Review. Russian.
-
RNA editing.Annu Rev Neurosci. 1996;19:27-52. doi: 10.1146/annurev.ne.19.030196.000331. Annu Rev Neurosci. 1996. PMID: 8833435 Review.
Cited by
-
Editing site recognition and nucleotide insertion are separable processes in Physarum mitochondria.EMBO J. 2002 Nov 15;21(22):6154-61. doi: 10.1093/emboj/cdf610. EMBO J. 2002. PMID: 12426387 Free PMC article.
-
Electroporation of DNA into Physarum polycephalum Mitochondria: Effects on Transcription and RNA Editing in Isolated Organelles.Genes (Basel). 2016 Dec 14;7(12):128. doi: 10.3390/genes7120128. Genes (Basel). 2016. PMID: 27983641 Free PMC article.
-
Distinct roles for sequences upstream of and downstream from Physarum editing sites.RNA. 2009 Sep;15(9):1753-65. doi: 10.1261/rna.1668309. Epub 2009 Jul 15. RNA. 2009. PMID: 19605532 Free PMC article.
-
Non-templated addition of nucleotides to the 3' end of nascent RNA during RNA editing in Physarum.EMBO J. 2001 Mar 15;20(6):1405-14. doi: 10.1093/emboj/20.6.1405. EMBO J. 2001. PMID: 11250906 Free PMC article.
-
Unexpectedly complex editing patterns at dinucleotide insertion sites in Physarum mitochondria.Mol Cell Biol. 2004 Sep;24(18):7821-8. doi: 10.1128/MCB.24.18.7821-7828.2004. Mol Cell Biol. 2004. PMID: 15340046 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

