Recognition and incision of site-specifically modified C8 guanine adducts formed by 2-aminofluorene, N-acetyl-2-aminofluorene and 1-nitropyrene by UvrABC nuclease
- PMID: 11000263
- PMCID: PMC110764
- DOI: 10.1093/nar/28.19.3719
Recognition and incision of site-specifically modified C8 guanine adducts formed by 2-aminofluorene, N-acetyl-2-aminofluorene and 1-nitropyrene by UvrABC nuclease
Abstract
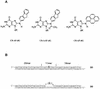
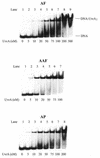
Nucleotide excision repair plays a crucial role in removing many types of DNA adducts formed by UV light and chemical carcinogens. We have examined the interactions of Escherichia coli UvrABC nuclease proteins with three site-specific C8 guanine adducts formed by the carcinogens 2-aminofluorene (AF), N:-acetyl-2-acetylaminofluorene (AAF) and 1-nitropyrene (1-NP) in a 50mer oligonucleotide. Similar to the AF and AAF adducts, the 1-NP-induced DNA adduct contains an aminopyrene (AP) moiety covalently linked to the C8 position of guanine. The dissociation constants for UvrA binding to AF-, AAF- and AP-DNA adducts, determined by gel mobility shift assay, are 33 +/- 9, 8 +/- 2 and 23 +/- 9 nM, respectively, indicating that the AAF adduct is recognized much more efficiently than the other two. Incision by UvrABC nuclease showed that AAF-DNA was cleaved approximately 2-fold more efficiently than AF- or AP-DNA (AAF > AF approximately AP), even though AP has the largest molecular size in this group. However, an opened DNA structure of six bases around the adduct increased the incision efficiency for AF-DNA (but not for AP-DNA), making it equivalent to that for AAF-DNA. These results are consistent with a model in which DNA damage recognition by the E. coli nucleotide excision repair system consists of two sequential steps. It includes recognition of helical distortion in duplex DNA followed by recognition of the type of nucleotide chemical modification in a single-stranded region. The difference in incision efficiency between AF- and AAF-DNA adducts in normal DNA sequence, therefore, is a consequence of their difference in inducing structural distortions in DNA. The results of this study are discussed in the light of NMR solution structures of these DNA adducts.
Figures




Similar articles
-
Recognition and incision of gamma-radiation-induced cross-linked guanine-thymine tandem lesion G[8,5-Me]T by UvrABC nuclease.Chem Res Toxicol. 2005 Sep;18(9):1339-46. doi: 10.1021/tx050147+. Chem Res Toxicol. 2005. PMID: 16167825 Free PMC article.
-
Differential incision of bulky carcinogen-DNA adducts by the UvrABC nuclease: comparison of incision rates and the interactions of Uvr subunits with lesions of different structures.Biochemistry. 2000 Oct 10;39(40):12252-61. doi: 10.1021/bi0013187. Biochemistry. 2000. PMID: 11015204
-
Rate of incision of N-acetyl-2-aminofluorene and N-2-aminofluorene adducts by UvrABC nuclease is adduct- and sequence-specific: comparison of the rates of UvrABC nuclease incision and protein-DNA complex formation.Biochemistry. 1998 Jan 13;37(2):571-9. doi: 10.1021/bi971544p. Biochemistry. 1998. PMID: 9425079
-
Recognition and repair of DNA-cisplatin adducts.Acta Biochim Pol. 2002;49(3):583-96. Acta Biochim Pol. 2002. PMID: 12422229 Review.
-
The formation and biological significance of N7-guanine adducts.Mutat Res. 2009 Aug;678(2):76-94. doi: 10.1016/j.mrgentox.2009.05.006. Epub 2009 May 22. Mutat Res. 2009. PMID: 19465146 Free PMC article. Review.
Cited by
-
Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair.Proc Natl Acad Sci U S A. 2002 Feb 19;99(4):1905-9. doi: 10.1073/pnas.042700399. Epub 2002 Feb 12. Proc Natl Acad Sci U S A. 2002. PMID: 11842222 Free PMC article.
-
Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease.Biochemistry. 2003 Nov 4;42(43):12654-61. doi: 10.1021/bi034446e. Biochemistry. 2003. PMID: 14580212 Free PMC article.
-
Surviving the sun: repair and bypass of DNA UV lesions.Protein Sci. 2011 Nov;20(11):1781-9. doi: 10.1002/pro.723. Protein Sci. 2011. PMID: 21898645 Free PMC article. Review.
-
Cooperative interaction of human XPA stabilizes and enhances specific binding of XPA to DNA damage.Biochemistry. 2005 May 17;44(19):7361-8. doi: 10.1021/bi047598y. Biochemistry. 2005. PMID: 15882075 Free PMC article.
-
(5'S)-8,5'-cyclo-2'-deoxyguanosine is a strong block to replication, a potent pol V-dependent mutagenic lesion, and is inefficiently repaired in Escherichia coli.Biochemistry. 2011 May 17;50(19):3862-5. doi: 10.1021/bi2004944. Epub 2011 Apr 25. Biochemistry. 2011. PMID: 21491964 Free PMC article.
References
-
- Rosenkranz H.S., McCoy,E.C., Sanders,D.R., Butler,M., Kiriazides,D.K. and Mermelstein,R. (1980) Science, 209, 1039–1043. - PubMed
-
- Tokiwa H. and Ohnishi,Y. (1986) CRC Crit. Rev. Toxicol., 17, 23–60. - PubMed
-
- IARC (1989) IARC Monographs on the Evaluation of Carcinogenic Risk to Humans, Vol. 39. IARC, Lyon.
-
- Kriek E. (1992) J. Cancer Res. Clin. Oncol., 118, 481–489. - PubMed
-
- Heflich R.H. and Neft,R.E. (1994) Mutat. Res., 318, 73–114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

