Photoaffinity polyamines: interactions with AcPhe-tRNA free in solution or bound at the P-site of Escherichia coli ribosomes
- PMID: 11000265
- PMCID: PMC110758
- DOI: 10.1093/nar/28.19.3733
Photoaffinity polyamines: interactions with AcPhe-tRNA free in solution or bound at the P-site of Escherichia coli ribosomes
Abstract
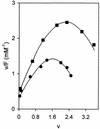
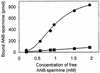
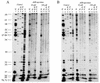
Two photoreactive derivatives of spermine, azidobenzamidino (ABA)-spermine and azidonitrobenzoyl (ANB)-spermine, were used for mapping of polyamine binding sites in AcPhe-tRNA free in solution or bound at the P-site of Escherichia coli poly(U)-programmed ribosomes. Partial nuclease digestion indicated that the deep pocket formed by nucleosides of the D-stem and the variable loop, as well as the anticodon stem, are preferable polyamine binding sites for AcPhe-tRNA in the free state. ABA-spermine was a stronger cross-linker than ANB-spermine. Both photoprobes were linked to AcPhe-tRNA with higher affinity when the latter was non-enzymatically bound to poly(U)-programmed ribosomes. In particular, the cross-linking at the TpsiC stem and acceptor stem was substantially promoted. The photolabeled AcPhe-tRNA.poly(U).ribosome complex exhibited moderate reactivity towards puromycin. The attachment of photoprobes to AcPhe-tRNA was mainly responsible for this defect. A more complicated situation was revealed when the AcPhe-tRNA.poly(U).ribosome complex was formed in the presence of translation factors; the reactivity towards puromycin was stimulated by irradiating such a complex in the presence of photoprobes at 50 microM, with higher concentrations being inhibitory. The stimulatory effect was closely related with the binding of photoprobes to ribosomes. The results are discussed on the basis of possible AcPhe-tRNA conformational changes induced by the incorporation of photoprobes.
Figures





Similar articles
-
Localization of spermine binding sites in 23S rRNA by photoaffinity labeling: parsing the spermine contribution to ribosomal 50S subunit functions.Nucleic Acids Res. 2005 May 16;33(9):2792-805. doi: 10.1093/nar/gki557. Print 2005. Nucleic Acids Res. 2005. PMID: 15897324 Free PMC article.
-
Changes in the conformation of 5S rRNA cause alterations in principal functions of the ribosomal nanomachine.Nucleic Acids Res. 2007;35(15):5108-19. doi: 10.1093/nar/gkm546. Epub 2007 Jul 25. Nucleic Acids Res. 2007. PMID: 17652323 Free PMC article.
-
Effects of two photoreactive spermine analogues on peptide bond formation and their application for labeling proteins in Escherichia coli functional ribosomal complexes.Biochemistry. 2001 Jun 26;40(25):7641-50. doi: 10.1021/bi010010s. Biochemistry. 2001. PMID: 11412118
-
[tRNA-binding centers of Escherichia coli ribosomes and their structural organization].Mol Biol (Mosk). 1984 Sep-Oct;18(5):1194-207. Mol Biol (Mosk). 1984. PMID: 6209546 Review. Russian.
-
[The effect of modification of tRNA nucleotide-37 on the tRNA interaction with the P- and A-site of the 70S ribosome Escherichia coli].Mol Biol (Mosk). 2006 Jul-Aug;40(4):669-83. Mol Biol (Mosk). 2006. PMID: 16913226 Review. Russian.
Cited by
-
Localization of spermine binding sites in 23S rRNA by photoaffinity labeling: parsing the spermine contribution to ribosomal 50S subunit functions.Nucleic Acids Res. 2005 May 16;33(9):2792-805. doi: 10.1093/nar/gki557. Print 2005. Nucleic Acids Res. 2005. PMID: 15897324 Free PMC article.
-
Probing tRNA interaction with biogenic polyamines.RNA. 2010 Oct;16(10):1968-79. doi: 10.1261/rna.1994310. Epub 2010 Aug 20. RNA. 2010. PMID: 20729276 Free PMC article.
-
Translational recoding as a feedback controller: systems approaches reveal polyamine-specific effects on the antizyme ribosomal frameshift.Nucleic Acids Res. 2011 Jun;39(11):4587-97. doi: 10.1093/nar/gkq1349. Epub 2011 Feb 7. Nucleic Acids Res. 2011. PMID: 21303766 Free PMC article.
-
Changes in the conformation of 5S rRNA cause alterations in principal functions of the ribosomal nanomachine.Nucleic Acids Res. 2007;35(15):5108-19. doi: 10.1093/nar/gkm546. Epub 2007 Jul 25. Nucleic Acids Res. 2007. PMID: 17652323 Free PMC article.
-
The identification of spermine binding sites in 16S rRNA allows interpretation of the spermine effect on ribosomal 30S subunit functions.Nucleic Acids Res. 2002 Jul 1;30(13):2832-43. doi: 10.1093/nar/gkf404. Nucleic Acids Res. 2002. PMID: 12087167 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

