Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1
- PMID: 11003647
- PMCID: PMC86303
- DOI: 10.1128/MCB.20.20.7505-7515.2000
Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1
Abstract
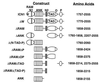
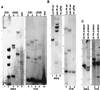
Notch receptors participate in a conserved signaling pathway that controls the development of diverse tissues and cell types, including lymphoid cells. Signaling is normally initiated through one or more ligand-mediated proteolytic cleavages that permit nuclear translocation of the intracellular portion of the Notch receptor (ICN), which then binds and activates transcription factors of the Su(H)/CBF1 family. Several mammalian Notch receptors are oncogenic when constitutively active, including Notch1, a gene initially identified based on its involvement in a (7;9) chromosomal translocation found in sporadic T-cell lymphoblastic leukemias and lymphomas (T-ALL). To investigate which portions of ICN1 contribute to transformation, we performed a structure-transformation analysis using a robust murine bone marrow reconstitution assay. Both the ankyrin repeat and C-terminal transactivation domains were required for T-cell leukemogenesis, whereas the N-terminal RAM domain and a C-terminal domain that includes a PEST sequence were nonessential. Induction of T-ALL correlated with the transactivation activity of each Notch1 polypeptide when fused to the DNA-binding domain of GAL4, with the exception of polypeptides deleted of the ankyrin repeats, which lacked transforming activity while retaining strong transactivation activity. Transforming polypeptides also demonstrated moderate to strong activation of the Su(H)/CBF1-sensitive HES-1 promoter, while polypeptides with weak or absent activity on this promoter failed to cause leukemia. These experiments define a minimal transforming region for Notch1 in T-cell progenitors and suggest that leukemogenic signaling involves recruitment of transcriptional coactivators to ICN1 nuclear complexes.
Figures




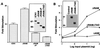
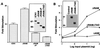


References
-
- Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Aster J, Pear W, Hasserjian R, Erba H, Davi F, Luo B, Scott M, Baltimore D, Sklar J. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harbor Symp Quant Biol. 1994;59:125–136. - PubMed
-
- Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. - PubMed
-
- Bailey A M, Posakony J W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
