Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators
- PMID: 11003651
- PMCID: PMC86307
- DOI: 10.1128/MCB.20.20.7550-7558.2000
Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators
Abstract
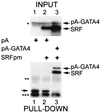
Combinatorial interaction among cardiac tissue-restricted enriched transcription factors may facilitate the expression of cardiac tissue-restricted genes. Here we show that the MADS box factor serum response factor (SRF) cooperates with the zinc finger protein GATA-4 to synergistically activate numerous myogenic and nonmyogenic serum response element (SRE)-dependent promoters in CV1 fibroblasts. In the absence of GATA binding sites, synergistic activation depends on binding of SRF to the proximal CArG box sequence in the cardiac and skeletal alpha-actin promoter. GATA-4's C-terminal activation domain is obligatory for synergistic coactivation with SRF, and its N-terminal domain and first zinc finger are inhibitory. SRF and GATA-4 physically associate both in vivo and in vitro through their MADS box and the second zinc finger domains as determined by protein A pullout assays and by in vivo one-hybrid transfection assays using Gal4 fusion proteins. Other cardiovascular tissue-restricted GATA factors, such as GATA-5 and GATA-6, were equivalent to GATA-4 in coactivating SRE-dependent targets. Thus, interaction between the MADS box and C4 zinc finger proteins, a novel regulatory paradigm, mediates activation of SRF-dependent gene expression.
Figures



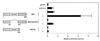


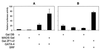
References
-
- Belaguli N S, Schildmeyer L A, Schwartz R J. Organization and myogenic restricted expression of the murine serum response factor gene. J Biol Chem. 1997;272:18222–18231. - PubMed
-
- Black B L, Olson E N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
