The last RNA-binding repeat of the Escherichia coli ribosomal protein S1 is specifically involved in autogenous control
- PMID: 11004188
- PMCID: PMC94711
- DOI: 10.1128/JB.182.20.5872-5879.2000
The last RNA-binding repeat of the Escherichia coli ribosomal protein S1 is specifically involved in autogenous control
Abstract
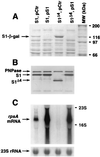
The ssyF29 mutation, originally selected as an extragenic suppressor of a protein export defect, has been mapped within the rpsA gene encoding ribosomal protein S1. Here, we examine the nature of this mutation and its effect on translation. Sequencing of the rpsA gene from the ssyF mutant has revealed that, due to an IS10R insertion, its product lacks the last 92 residues of the wild-type S1 protein corresponding to one of the four homologous repeats of the RNA-binding domain. To investigate how this truncation affects translation, we have created two series of Escherichia coli strains (rpsA(+) and ssyF) bearing various translation initiation regions (TIRs) fused to the chromosomal lacZ gene. Using a beta-galactosidase assay, we show that none of these TIRs differ in activity between ssyF and rpsA(+) cells, except for the rpsA TIR: the latter is stimulated threefold in ssyF cells, provided it retains at least ca. 90 nucleotides upstream of the start codon. Similarly, the activity of this TIR can be severely repressed in trans by excess S1, again provided it retains the same minimal upstream sequence. Thus, the ssyF stimulation requires the presence of the rpsA translational autogenous operator. As an interpretation, we propose that the ssyF mutation relieves the residual repression caused by normal supply of S1 (i.e., that it impairs autogenous control). Thus, the C-terminal repeat of the S1 RNA-binding domain appears to be required for autoregulation, but not for overall mRNA recognition.
Figures





References
-
- Artamonova V S, Boni I V. The ssyF29 mutation in E. coli ribosomal protein S1 gene which suppresses the defect in protein export machinery results from an insertion of IS10R-element. Bioorg Khim. 1996;22:941–943. - PubMed
-
- Artamonova V S, Tsareva N V, Boni I V. Regulation of the ribosomal protein L7/12 synthesis: the role of the rplJL intercistronic region as a translational enhancer. Bioorg Khim. 1998;24:530–538. - PubMed
-
- Boni I V, Zlatkin I V, Budowsky E I. Ribosomal protein S1 associates with Escherichia coli ribosomal 30S subunit by means of protein-protein interactions. Eur J Biochem. 1982;121:371–376. - PubMed
-
- Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

