Characterization of the mouse gene for the heavy metal-responsive transcription factor MTF-1
- PMID: 11005378
- PMCID: PMC312886
- DOI: 10.1379/1466-1268(2000)005<0196:cotmgf>2.0.co;2
Characterization of the mouse gene for the heavy metal-responsive transcription factor MTF-1
Abstract
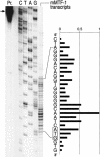
MTF-1 is a zinc finger transcription factor that mediates the cellular response to heavy metal stress; its targeted disruption in the mouse leads to liver decay and embryonic lethality at day E14. Recently, we have sequenced the entire MTF-1 gene in the compact genome of the pufferfish Fugu rubripes. Here we have defined the promoter sequences of human and mouse MTF-1 and the genomic structure of the mouse MTF-1 locus. The transcription unit of MTF-1 spans 42 kb (compared to 8.5 kb in Fugu) and is located downstream of the gene for a phosphatase (INPP5P) in mouse, human, and fish. In all of these species, the MTF promoter region has the features of a CpG island. In both mouse and human, the 5' untranslated region harbors conserved short reading frames of unknown function. RNA mapping experiments revealed that in these two species, MTF-1 mRNA is transcribed from a cluster of multiple initiation sites from a TATA-less promoter without metal-responsive elements. Transcription from endogenous and transfected MTF-1 promoters was not affected by heavy metal load or other stressors, in support of the notion that MTF-1 activity is regulated at the posttranscriptional level. Tissue Northern blots normalized for poly A+ RNA indicate that MTF-1 is expressed at similar levels in all tissues, except in the testes, that contain more than 10-fold higher mRNA levels.
Figures




References
-
- Auf der Maur A, Belser T, Elgar G, Georgiev O, Schaffner W. Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of heavy metal stress response. Biol Chem. 1999;380:175–185. - PubMed
-
- Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NK-kappa B. Nature. 1995;376:167–170. - PubMed
-
- Bittel D, Dalton T, Samson SL, Gedamu L, Andrews GK. The DNA binding activity of metal response element-binding transcription factor-1 is activated in vivo and in vitro by zinc, but not by other transition metals. J Biol Chem. 1998;273:7127–7133. - PubMed
-
- Brown CY, Mize GJ, Pineda M, George DL, Morris DR. Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene. 1999;18:5631–5637. - PubMed
-
- Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
