Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions
- PMID: 11005831
- PMCID: PMC17252
- DOI: 10.1073/pnas.190309197
Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions
Abstract
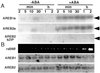
The induction of the dehydration-responsive Arabidopsis gene, rd29B, is mediated mainly by abscisic acid (ABA). Promoter analysis of rd29B indicated that two ABA-responsive elements (ABREs) are required for the dehydration-responsive expression of rd29B as cis-acting elements. Three cDNAs encoding basic leucine zipper (bZIP)-type ABRE-binding proteins were isolated by using the yeast one-hybrid system and were designated AREB1, AREB2, and AREB3 (ABA-responsive element binding protein). Transcription of the AREB1 and AREB2 genes is up-regulated by drought, NaCl, and ABA treatment in vegetative tissues. In a transient transactivation experiment using Arabidopsis leaf protoplasts, both the AREB1 and AREB2 proteins activated transcription of a reporter gene driven by ABRE. AREB1 and AREB2 required ABA for their activation, because their transactivation activities were repressed in aba2 and abi1 mutants and enhanced in an era1 mutant. Activation of AREBs by ABA was suppressed by protein kinase inhibitors. These results suggest that both AREB1 and AREB2 function as transcriptional activators in the ABA-inducible expression of rd29B, and further that ABA-dependent posttranscriptional activation of AREB1 and AREB2, probably by phosphorylation, is necessary for their maximum activation by ABA. Using cultured Arabidopsis cells, we demonstrated that a specific ABA-activated protein kinase of 42-kDa phosphorylated conserved N-terminal regions in the AREB proteins.
Figures



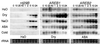

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

