Remarkable species diversity in Malagasy mouse lemurs (primates, Microcebus)
- PMID: 11005834
- PMCID: PMC17199
- DOI: 10.1073/pnas.200121897
Remarkable species diversity in Malagasy mouse lemurs (primates, Microcebus)
Abstract
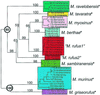
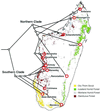
Phylogenetic analysis of mtDNA sequence data confirms the observation that species diversity in the world's smallest living primate (genus Microcebus) has been greatly underestimated. The description of three species new to science, and the resurrection of two others from synonymy, has been justified on morphological grounds and is supported by evidence of reproductive isolation in sympatry. This taxonomic revision doubles the number of recognized mouse lemur species. The molecular data and phylogenetic analyses presented here verify the revision and add a historical framework for understanding mouse lemur species diversity. Phylogenetic analysis revises established hypotheses of ecogeographic constraint for the maintenance of species boundaries in these endemic Malagasy primates. Mouse lemur clades also show conspicuous patterns of regional endemism, thereby emphasizing the threat of local deforestation to Madagascar's unique biodiversity.
Figures
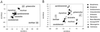

References
-
- Coyne J A, Orr H A. In: Speciation and Its Consequences. Otte D, Endler J A, editors. Sunderland, MA: Sinauer; 1989. pp. 180–207.
-
- Cracraft J. Curr Ornithol. 1983;1:159–187.
-
- Davis J I, Nixon K C. Syst Biol. 1992;41:421–435.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

