Identification of a third distinct estrogen receptor and reclassification of estrogen receptors in teleosts
- PMID: 11005855
- PMCID: PMC27095
- DOI: 10.1073/pnas.97.20.10751
Identification of a third distinct estrogen receptor and reclassification of estrogen receptors in teleosts
Abstract
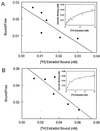
This paper describes three distinct estrogen receptor (ER) subtypes: ERalpha, ERbeta, and a unique type, ERgamma, cloned from a teleost fish, the Atlantic croaker Micropogonias undulatus; the first identification of a third type of classical ER in vertebrate species. Phylogenetic analysis shows that ERgamma arose through gene duplication from ERbeta early in the teleost lineage and indicates that ERgamma is present in other teleosts, although it has not been recognized as such. The Atlantic croaker ERgamma shows amino acid differences in regions important for ligand binding and receptor activation that are conserved in all other ERgammas. The three ER subtypes are genetically distinct and have different distribution patterns in Atlantic croaker tissues. In addition, ERbeta and ERgamma fusion proteins can each bind estradiol-17beta with high affinity. The presence of three functional ERs in one species expands the role of ER multiplicity in estrogen signaling systems and provides a unique opportunity to investigate the dynamics and mechanisms of ER evolution.
Figures

 ) (23). Positions that contact ligands in
mammalian ERs are noted with an asterisk (24, 25). The amino acids in
the LBD known to be critical for E2 binding are noted with arrows (26).
Sequence notation includes alignment gaps (–) and identity with
croaker ER gamma (.).
) (23). Positions that contact ligands in
mammalian ERs are noted with an asterisk (24, 25). The amino acids in
the LBD known to be critical for E2 binding are noted with arrows (26).
Sequence notation includes alignment gaps (–) and identity with
croaker ER gamma (.).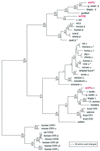


References
-
- Kuiper G G J M, Carlsson B, Grandien K, Enmark E, Hagglad J, Nilsson S, Gustafsson J-Å. Endocrinology. 1997;138:863–870. - PubMed
-
- Paech K, Webb P, Kuiper G G J M, Nilsson S, Gustafsson J-Å, Kushner P J, Scanlan T S. Science. 1997;277:1508–1510. - PubMed
-
- Todo T, Adachi S, Yamauchi K. Mol Cell Endocrinol. 1996;119:37–45. - PubMed
-
- Mosselman S, Polman J, Dijkema R. FEBS Lett. 1996;392:49–53. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

