Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination
- PMID: 11005857
- PMCID: PMC27106
- DOI: 10.1073/pnas.97.20.10814
Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination
Abstract
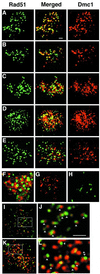
Two RecA homologs, Rad51 and Dmc1, assemble as cytologically visible complexes (foci) at the same sites on meiotic chromosomes. Time course analysis confirms that co-foci appear and disappear as the single predominant form. A large fraction of co-foci are eliminated in a red1 mutant, which is expected as a characteristic of the interhomolog-specific recombination pathway. Previous studies suggested that normal Dmc1 loading depends on Rad51. We show here that a mutation in TID1/RDH54, encoding a RAD54 homolog, reduces Rad51-Dmc1 colocalization relative to WT. A rad54 mutation, in contrast, has relatively little effect on RecA homolog foci except when strains also contain a tid1/rdh54 mutation. The role of Tid1/Rdh54 in coordinating RecA homolog assembly may be very direct, because Tid1/Rdh54 is known to physically bind both Dmc1 and Rad51. Also, Dmc1 foci appear early in a tid1/rdh54 mutant. Thus, Tid1 may normally act with Rad51 to promote ordered RecA homolog assembly by blocking Dmc1 until Rad51 is present. Finally, whereas double-staining foci predominate in WT nuclei, a subset of nuclei with expanded chromatin exhibit individual Rad51 and Dmc1 foci side-by-side, suggesting that a Rad51 homo-oligomer and a Dmc1 homo-oligomer assemble next to one another at the site of a single double-strand break (DSB) recombination intermediate.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

